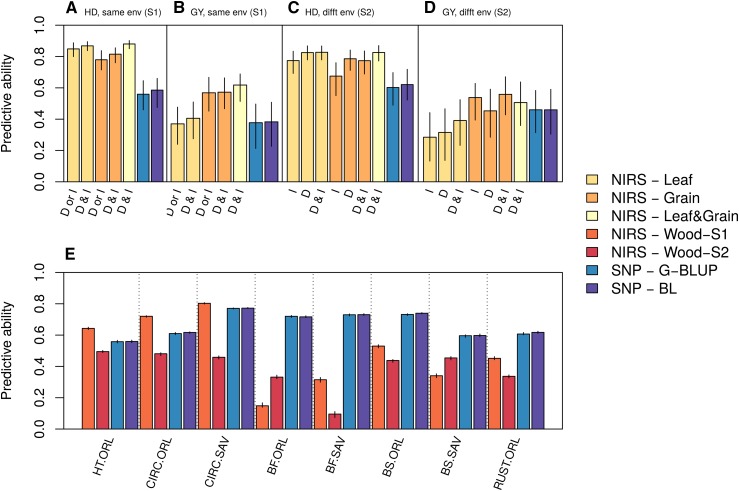
Figure 3.
Predictive ability of SNP (G-BLUP or Bayesian LASSO (BL) models) or NIRS (RR-BLUP model) when predicting the phenotypic values of individuals within a cross-validation in winter wheat (a, b, c, d) and black poplar (e). Two scenarios were considered for NIRS prediction: in S1, the RR-BLUP model was trained with NIRS data and phenotypes that were collected within the same environment (a, b, e), whereas in S2, NIRS and phenotypic data used to train the RR-BLUP model were collected in distinct environments (c, d, e). For wheat, two traits were considered: heading date (a, c) and grain yield (b, d). The bars of a, b, c, and d are labeled with the origin of the NIRS data (I: irrigated treatment, D: drought treatment), and the bars of e are labeled with the combination of trait and experiment (HT: height, CIRC: circumference, BF: bud flush, BS: bud set, RUST: resistance to rust, ORL: experimental design in Orléans, France, SAV: experimental design in Savigliano, Italy). For S1 in wheat (a, b), PS was done with NIRS collected in the environment in which the phenotypes were collected (D or I), or with NIRS collected in both environments (D & I). For S2 in wheat, each boxplot represents the summary statistics over all the accuracies obtained in the six S2 environments with a given type of NIRS (tissue and environment). The medians of the accuracies obtained over repeated cross-validations are reported as the height of the bars together with the first and third quartiles as confidence intervals.