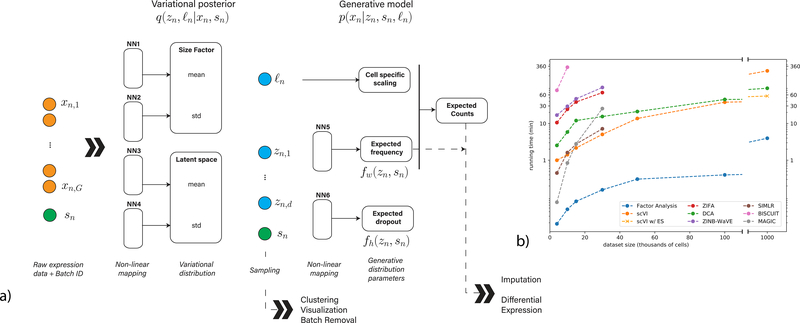
Figure 1:
Overview of scVI. Given a gene-expression matrix with batch annotations as input, scVI learns a non-linear embedding of the cells that can be used for multiple analysis tasks. (a) The computational trees (neural networks) used to compute the embedding as well as the distribution of gene expression. (b) Comparison of running times (y-axis) on the BRAIN-LARGE data with a limited set of 720 genes, and with increasing input sizes (x-axis; cells in each input set are sampled randomly from the complete dataset). All the algorithms were tested on a machine with one eight-core Intel i7–6820HQ CPU addressing 32 GB RAM, and one NVIDIA Tesla K80 (GK210GL) GPU addressing 24 GB RAM. scVI is compared against existing methods for dimensionality reduction in the scRNA-seq literature. As a control, we also add basic matrix factorization with factor analysis (FA). For the one-million-cell dataset only, we report the result of scVI with and without early stopping (ES).