Fig. 1.
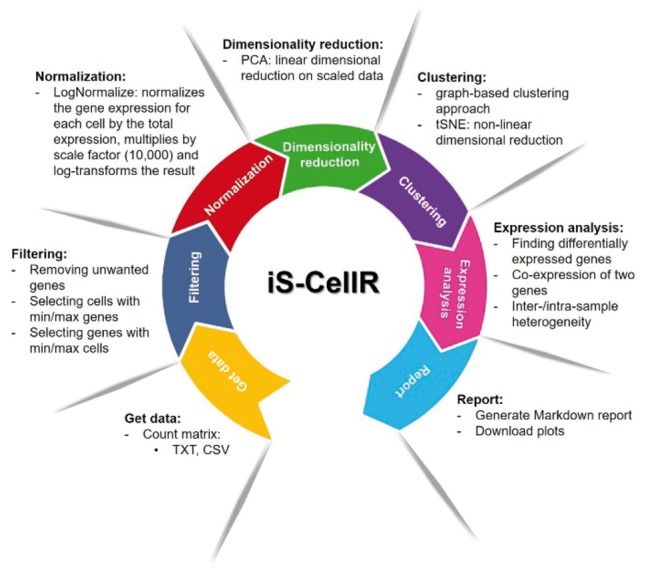
iS-CellR pipeline overview. iS-CellR is organized into a seven-step process for complete scRNA-seq analysis. The user can interactively select steps to perform analysis using single-cell data. After uploading, the raw data are filtered and normalized. The normalized data are then subjected to dimensionality reduction for principle component analysis (PCA). Further dimensionality reduction can be performed using t-distributed stochastic neighbour embedding (tSNE). After a clustering step, differentially expressed marker genes can be visualized on cell clusters. The user can also visualize co-expression of two genes simultaneously. Inter-/intra-sample heterogeneity requires the user to upload a file with a list of genes in a two-column format (GeneSet1, GeneSet2). Finally, the user can generate a HTML report containing all results produced or download plots individually
