Fig. 1.
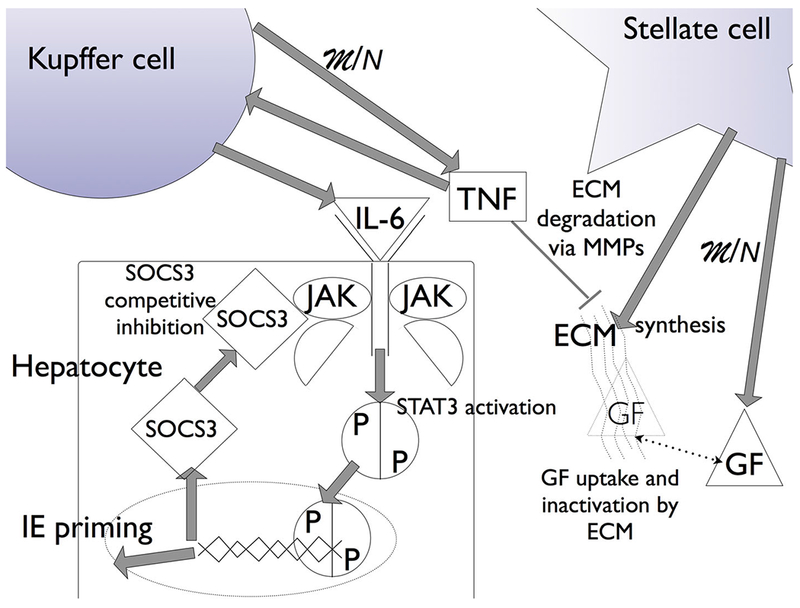
Liver regeneration biochemistry in the model. The increase in metabolic load (M) due to a reduction in hepatocyte number (N) drives hepatocyte growth factor (GF) expression and tumor necrosis factor (TNF) expression, from stellate and Kupffer cells, respectively. TNF initiates the degradation of the ECM via matrix metalloproteinases (MMPs) and the expression of cytokines (IL-6) that initiate janus kinase (JAK) signaling through STAT3 activation. STAT3 translocation into the nucleus leads to the expression of suppressor of cytokine signaling 3 (SOCS3), and the expression of immediate early genes (IE) that signal quiescent hepatocytes to enter a primed state. The competitive binding of SOCS3 to JAK leads to suppression of STAT3 signaling, providing an activating pulse. GF drives primed cells to a proliferating state. As the metabolic load decreases due to the increase in hepatocyte number, TNF concentration decreases and the ECM reforms, and GF uptake by the ECM leads to a cessation in primed cells moving to proliferation. The ECM reforming also drives proliferating cells to quiescence, bringing liver regeneration to a halt.
