Abstract
Recent advances in direct imaging have given us a new appreciation of the spatial and temporal dynamics of membrane trafficking processes, and have allowed us to ask questions that were difficult to address with traditional methods. A relevant example of this is protein sorting in the endosome, which serves as the primary sorting station for proteins internalized from the cell surface. In this chapter, we discuss fluorescence imaging protocols to directly visualize and quantitate the recycling of G protein-coupled receptors (GPCRs)—a highly physiologically relevant family of signaling receptors—in real time in living cells. The protocols allow direct visualization and quantitation of both GPCR exit from the endosome and GPCR delivery to the cell surface. The methods may be extended to study the endolysosomal sorting of many proteins that undergoes endocytic cycling, and may be adapted to other organelles and systems where proteins are sorted.
INTRODUCTION
How organelles sort cargo proteins to various destinations while maintaining their own identity is a fundamental question in cell biology. The early endosome, the primary sorting station for internalized proteins, serves as a poster child for this question. The endosome sorts internalized membrane proteins to one of four main pathways. First, nutrient receptors are transported to the cell surface as part of bulk membrane flow by geometric sorting (Dunn, McGraw, & Maxfield, 1989; Maxfield & McGraw, 2004; Mayor, Presley, & Maxfield, 1993). Second, many signaling receptors are targeted to the lysosome to be degraded (Hislop, Henry, & von Zastrow, 2011; Huang et al., 2013; Marchese, Paing, Temple, & Trejo, 2008; Tanowitz & Von Zastrow, 2002; Trejo, 1999). Third, some proteins are transported in a retrograde manner to the Golgi apparatus (Cheng & Filardo, 2012; Chia, Gunn, & Gleeson, 2013; Johannes & Wunder, 2011; Zhao et al., 2013). Fourth, many members of the G protein-coupled receptor (GPCR) family—by far the largest and physiologically most diverse family of signaling receptors—are recycled in a regulated sequence-dependent manner, distinct from bulk recycling (Hanyaloglu & von Zastrow, 2008).
While the mechanisms directing proteins into the first three pathways (geometric sorting by tubule-forming proteins, involution by the endosomal sorting complexes required for transport proteins, and retrograde transport by the retromer complex) have been studied at structural and functional levels, we have only recently made inroads into understanding sequence-dependent recycling of GPCRs (Drake, Shenoy, & Lefkowitz, 2006; Hanyaloglu & von Zastrow, 2008; Marchese et al., 2008; Romero, von Zastrow, & Friedman, 2011). A major hurdle in understanding GPCR sorting had been the lack of assays to directly visualize and quantitate endosomal sorting in living cells in real time. Recent advances in fluorescence microscopy-based approaches have enabled us to overcome this hurdle and to resolve endosomal sorting with high spatial and temporal resolution (Puthenveedu et al., 2010; Vistein & Puthenveedu, 2013; Yu, Dhavan, Chevalier, Yudowski, & Zastrow, 2010; Yudowski, Puthenveedu, Henry, & von Zastrow, 2009). A relevant aspect of this is that small kinetic changes in recycling, as is seen with physiological regulation by signaling pathways, are often difficult to detect using traditional assays, but these might cause fairly significant cumulative changes at physiological timescales.
Here we describe protocols for using fluorescence imaging to visualize and quantitate the recycling of internalized GPCRs, using two prototypic receptors—the beta 2 adrenergic receptor (B2AR) and the mu opioid receptor (MOR). We outline methods to image both the sorting of GPCRs into recycling domains on the endosome and their delivery to the cell surface, at ensemble (whole cell) scales as well as at single-event resolution. These assays, using live cell imaging in real time, allow for detection of even small changes in the rates of recycling.
1. OBJECTIVES AND RATIONALE
The overall goal of this chapter is to define protocols for quantitating GPCR recycling using fluorescence imaging. We will outline protocols using confocal microscopy and total internal reflection fluorescence microscopy (TIRFM) to measure receptor delivery to the cell surface at ensemble and single-event spatial scales. We will also describe the use of confocal microscopy to visualize the sorting of fluorescently tagged receptors at the endosome, spatially resolved at the level of individual microdomains on endosomes that mediate GPCR recycling.
To measure recycling at the whole cell level, receptors tagged to a pH-sensitive GFP (superecliptic pHluorin, SpH) will be used as a biosensor. This fluorophore is highly fluorescent at pH 7.1 or above. Reduction of pH below 7.1 causes a rapid single protonation of the GFP, and a practically instantaneous reduction in fluorescence by ~50-fold (Miesenböck, De Angelis, & Rothman, 1998). When receptors are tagged with SpH on their extracellular N-termini, the total fluorescence is an accurate estimate of the surface receptor levels, because the pH of most intracellular organelles is less than 7, and because endocytic vesicles are rapidly acidified (pH < 7). This property makes this sensor ideal to measure dynamic changes in surface receptor levels in response to activation. Endocytosis following receptor activation causes a fluorescence decrease because the SpH is in the lumen of endosomes, and recycling causes an increase in fluorescence, as the SpH is exposed to the extracellular medium and is dequenched when receptors return to the plasma membrane. Individual recycling events can be measured by imaging receptors tagged with SpH, using TIRFM to visualize the surface of cells. As vesicles of SpH-tagged receptors fuse with the membrane, SpH is dequenched and individual recycling events can be detected as transient bursts of fluorescence. To measure the dynamics of GPCR endosomal sorting, receptors N-terminally tagged with a FLAG epitope are specifically labeled on the cell surface with fluorescent anti-FLAG. Labeled receptors are visualized in endosomes, following endocytosis, using live cell confocal fluorescence microscopy with high spatial and temporal resolution.
2. MATERIALS AND INSTRUMENTS
2.1 REAGENTS
Human embryonic kidney (HEK 293) cells
Plasmid constructs encoding signal sequence (ss)-superecliptic pHluorin (SpH)-tagged GPCR or signal sequence (ss)-FLAG (DDDDK) epitope-tagged GPCR of choice (e.g., the MOR or B2AR)
Dulbecco’s minimum essential medium (DMEM), high glucose
Leibovitz’s L15, CO2-independent medium, without phenol red
Fetal bovine serum (FBS)
Effectene transfection reagent, Qiagen (or an equivalent transfection reagent)
Geneticin (G418)
#1.5 mm coverslip
Specific agonist for the GPCR—e.g., [D-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin (DAMGO) for the MOR or isoproterenol for the B2AR.
Immersion oil, type 37LDF (Cargille)
2.2 EQUIPMENT
A microscopy system is capable of imaging in the confocal and TIRFM modes, with appropriate excitation modes. We use a Nikon Eclipse Ti-inverted microscope with a 60× or 100× 1.49 NA. TIRF objective, with an Andor Revolution spinning disk confocal system, using 50 mW 488 and 561 DPSS lasers as light sources with appropriate filter sets. The microscope stage is housed within a temperature- and environment-controlled chamber.
Coverslip holder and chamber for live imaging. Alternatively, glass-bottomed dishes in which to plate cells.
3. METHODS
3.1 CELL CULTURE, PLATING, AND TRANSFECTION
HEK 293 cells are grown in DMEM with 10% by volume FBS at 37 °C with 5% CO2. Phenol red containing DMEM may be used during cell culture, but should be removed during any live cell microscopy assay as the phenol red contributes significantly to background fluorescence. Cells are plated onto #1.5 mm coverslip to reach 70% confluency after overnight incubation. The cells are transfected with SpH-tagged receptor (for the ensemble assay, Section 3.3.1, or the single-event recycling assay, Section 3.3.2) or Flag-tagged receptor (endosomal sorting assay, Section 3.3.3) using a lipid-based transfection reagent, such as Effectene (Qiagen). Cells should be imaged at 72–96 h after transfection. Alternatively, transfected cells may be placed under selection for 1–2 weeks to generate a stable cell line for the expressed plasmid. The stable cell lines are plated onto #1.5 coverslip for imaging at 50–90% confluency.
3.2 LIVE CELL IMAGING CONDITIONS
The coverslips with HEK 293 cells are moved to a live cell imaging chamber, and imaged in Leibovitz’s L15, CO2-independent medium, without phenol red and with 5% FBS (imaging media). For live cell imaging of receptor recycling and endosomal sorting, it is essential that experiments are performed at 37 °C, the physiological temperature for mammalian cells, because membrane trafficking events are very sensitive to temperature. Individual imaging protocols are described below for three separate experiments: ensemble measurement of surface levels, quantifying recycling at single-event resolution, and visualization of sorting and recycling in the endosome.
3.3 EXPERIMENTAL STRATEGIES
3.3.1 Strategy 1: ensemble measurement of surface levels
3.3.1.1 Background and objective
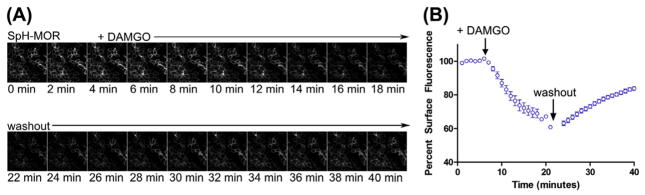
A straightforward method to assay GPCR recycling is to measure the overall number of receptors at the cell surface across several cells. Here, we describe an ensemble assay to measure the surface levels of receptors over time. Cells expressing the SpH–N-terminally tagged MORs are used as an example, described in Section 1. SpH-MOR will fluoresce at the cell surface when SpH is exposed to the neutral environment of the extracellular medium. When SpH-MORs are activated by the synthetic opioid peptide, DAMGO (described in Materials), endocytosis will redistribute MOR to acidic endosomal compartments, and SpH-MOR fluorescence will decrease as SpH is quenched (Figure 1(A)). This decrease can be used to measure endocytosis of receptors in a time-resolved manner. Similarly, recycling can be measured as an increase in surface fluorescence of SpH-MOR upon removal of agonist (Figure 1(A)). Cells are imaged at 20× magnification with a relatively low (0.4–0.75) numerical aperture objective, for optimum depth of field and optimum number of cells within one field. Several fields can be imaged to gather a large sample size of cells. The effects of drugs and other manipulations in the cell can be measured across treatments by adding drugs with agonist or during the washout to test for an effect on receptor recycling.
FIGURE 1. Strategy 1: Ensemble measurement of surface levels.
(A) Montage of fields visualized in the live cell ensemble SpH recycling assay. SpH-MOR is localized primarily to the surface of cells. Following DAMGO addition, surface fluorescence decreases over time as receptors endocytose and SpH is quenched in acidic endosomes. Following DAMGO washout, and addition of antagonist, SpH-MOR recycles, as seen by the increase of cell surface signal. (B) Example of SpH-MOR fluorescence intensity data in ensemble recycling assay. The mean fluorescence intensity was measured for a whole field of cells, and the graph shows the average fluorescence intensity across 20 fields. After DAMGO was added, SpH-MOR intensity decreased to around 50% of the baseline fluorescence after 20 min, indicative of receptor endocytosis. Following the washout, mean fluorescence increased to approximately 80% of the initial surface fluorescence after 20 min. Error bars are s.e.m.
3.3.1.2 Flow of experiments
Plate SpH-tagged receptor, for example, SpH-MOR, on #1.5 coverslip. We generate stable cell lines with Geneticin selection, following transfection of SpH-MOR with the Effectene transfection reagent, according to the manufacturer’s protocol.
Mount coverslips in a live cell imaging chamber. Alternatively, glass bottom dishes can be used.
Replace growth medium (i.e., DMEM + 10% FBS if using HEK 293 cells) with imaging medium (L15 with 5% FBS).
Use a lower magnification objective (i.e., 20x) in order to include several cells in one field.
Using confocal fluorescence microscopy and a 488 nm excitation laser, focus cells until membrane SpH fluorescence is crisp and concentrated at the periphery of the cell (Figure 1(A)). For a GPCR-like MOR that is expressed primarily at the cell surface, a strong surface SpH signal should be visible.
Choose 10 fields of cells with similar brightness and cell number. It is best if cells are 90–95% confluent and in a smooth monolayer.
Acquire a time-lapse image, acquiring every 1 min, with the lowest laser intensity that gives a reasonable dynamic range, to prevent phototoxicity and bleaching of SpH.
Acquire a 5 min baseline of surface SpH fluorescence. This will give an estimate of dynamic range and bleaching.
To measure recycling following a washout of receptor agonist, acquire a 15–20 min time lapse following agonist addition, then wash out agonist media two times, replace it with antagonist media, and acquire a 20–30 min time lapse to measure recycling.
To quantitate endocytosis and recycling, analyze the mean fluorescence intensity of each field of cells, or of each individual cell using image segmentation, and plot the mean fluorescence intensity over time.
3.3.1.3 Considerations
Persistent agonist exposure shifts the steady of receptors to an intracellular pool. Therefore, washing out agonist and replacing it with antagonist allow for detection of recycling without the confounding effects of continued endocytosis (Bowman et al., 2015; Hanyaloglu, McCullagh, & von Zastrow, 2005; Tanowitz & von Zastrow, 2003; Yu, Arttamangkul, Evans, Williams, & von Zastrow, 2009). However, rapid recycling begins shortly after agonist-induced endocytosis (Vistein & Puthenveedu, 2013; Yudowski et al., 2009). Therefore, it is important to consider that the SpH-tagged receptor levels in the presence of agonist are a combination of both recycling and endocytosis. A strategy to specifically measure recycling in the presence of persistent agonist is described below in Strategy 2—Quantifying recycling at single-event resolution.
3.3.1.4 Results
The fluorescence intensity over time will measure endocytosis and recycling of receptors during the time lapse. As receptors endocytose, the mean fluorescence intensity decreases, and as the receptors recycle back to the cell surface, it will increase. For example, SpH-MOR mean intensity decreases during the 20 min of agonist exposure, consistent with receptor endocytosis to endosomes and quenching of the SpH sensor (Figure 1(B)). Following the agonist washout, SpH-MOR fluorescence intensity increased, indicative of MOR recycling and dequenching of SpH as vesicles fuse with the plasma membrane (Figure 1(B)). The average changes of fluorescence typically fit single exponential curves, as expected.
3.3.2 Strategy 2: quantifying recycling at single-event resolution
3.3.2.1 Background and objective
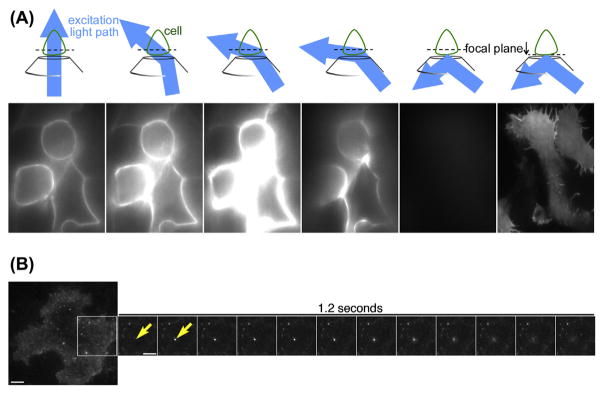
The surface population of receptors measured in the presence of agonist reflects the balance between endocytosis and recycling. Therefore, ensemble assays rely on experimental paradigms such as agonist washout to estimate recycling. In the presence of agonist, independent measures of endocytosis, measured under conditions known to block recycling, are needed for ensemble assays to indirectly estimate recycling. Here we describe the use of TIRFM to directly visualize individual vesicle fusion events that mediate GPCR recycling in real time in live cells. TIRFM relies on finding the critical angle where the light source does not pass through the sample, but instead gets reflected back toward the source (Ambrose, 1956; Axelrod, 1981). This specifically illuminates the cell surface nearest to the coverslip (Figure 2(A)). This assay can be used to directly resolve recycling without the confounding effects of endocytosis, even in the persistent presence of agonist (Vistein & Puthenveedu, 2013; Yudowski et al., 2009). This assay is very efficient for quantitating the changes in the number of recycling events per unit area per time, as well as the amount of receptors in individual recycling events, in the same cells after acute manipulations such as drug treatments. This minimizes cell to cell variability, and allows collection of statistically significant data from low number of cells.
FIGURE 2. Strategy 2: Quantifying recycling at single-event resolution.
(A) Schematic and corresponding example images for focusing a cell in TIRFM. When the excitation light passes straight through the sample, the center of the cell is readily visible. As the angle is moved down toward the critical angle, out of focus structures become more apparent in the “low-angle” illumination. Once past the critical angle, only the plasma membrane near the coverslip will be illuminated. The cell should appear bright, crisp, and uniform when focused down. (B) Example of a single recycling event. The whole cell is shown at the left. The inset focuses on a single recycling event. The recycling event first appears as a single bright spot whose fluorescence eventually diffuses out as vesicles fuse. Individual recycling events are very fast, each lasting less than a second. The scale bar is 5 μm. (See color plate)
3.3.2.2 Flow of experiment
Plate cells expressing SpH-tagged receptor on #1.5 mm coverslip so they are 50–90% confluent 48 h after plating, on the day of imaging.
Mount the coverslip onto the live cell imaging chamber in L15 medium containing 5% FBS by volume.
Add a drop of immersion oil on top of the oil immersion 1.49 NA 60× TIRF objective.
Place the live cell imaging chamber on the stage and bring cells into focus.
Switch to TIRFM illumination mode. To do this, first, focus on the center of the cell. Since the SpH tag is fluorescent practically only on the surface, the plasma membrane should appear as a ring (Figure 2(A), images on the left). Then move the angle of the laser down until all fluorescence is lost. Then focus down toward the coverslip to see the plasma membrane in the TIRF field (Figure 2(A), images on the right).
Select a bright (1000 counts or higher), large, flat cell that does not overlap with other cells. Nonoverlapping cells will be easier to analyze and quantify.
Activate the receptors by addition of agonist and allow for internalization for 5 min. The time may be adjusted depending on the response of the receptor.
Turn up the laser power to 60–70% to bleach the surface receptors until the signal events become evident. This will take about 10 s.
Image the cell for 1 min at 10 frames per second. Recycling events appear as abrupt bursts of fluorescence that rapidly dissipate.
Add secondary treatment to cells. This can be any acute manipulation, such as drugs.
Take second image for 1 min at 10 frames per second.
Save all image files. Have an unbiased third party or computer script scramble all file names before quantification.
Manually count the number of recycling events per cell by selecting a region of interest. Count each cell three times and take the average of these trials as the final recycling event count number.
Normalize the number of recycling events to the first series. If doing a non-paired comparison (i.e., comparing different cells) normalize the areas to compare the number of recycling events per unit area.
3.3.2.3 Considerations
Although the vast majority of events detected in the assay are recycling there is the potential to detect insertion of newly synthesized receptors from the biosynthetic pathway. Mock agonist treatment or cycloheximide pretreatment controls should be performed to verify that the fusion events are indeed recycling events, and not insertion of newly synthesized proteins. Recycling events require prior agonist-mediated internalization, and will not be affected by cycloheximide.
Due to the pH-sensitive nature of the SpH tag, the fluorescence should be restricted to the cell surface. Internal fluorescence may be caused by misfolded receptors or an imbalance of cellular pH. Unless there is constitutive recycling, there should be no clustering or puncta at the cell surface before agonist addition. Endocytosis receptor clusters are morphologically and temporally different. Recycling events are fast, on the order of about 1 s whereas endocytic clusters persist longer from 40 s to minutes long.
3.3.2.4 Results
When a cell is properly focused in TIRFM, the plasma membrane should be the only plane in focus (Figure 2(A), far right). Background fluorescence from parts of the cell not at the plasma membrane should disappear, and raising the focus away from the plasma membrane will not be possible and only yield a blurry, faint image (Figure 2(A)). To find the critical angle to make focusing in TIRFM possible, begin by focusing at the center of the cell (outside the TIRFM field). When the critical angle is reached this part of the cell will no longer be in focus. Figure 2(B) shows a time course of a single recycling event, within the context and scale of an entire cell. Recycling events are characterized by a sharp immediate increase in fluorescence, when the vesicle fuses with the plasma membrane, followed by a fast spreading of this fluorescence as the receptors diffuse on the plasma membrane. These events are typically very fast, lasting around 1 s. The particular example shown lasts 1.2 s.
3.3.3 Strategy 3: visualization of sorting and recycling in the endosome
3.3.3.1 Background and objective
GPCRs are sorted into cell surface recycling pathways at the early endosome. In addition to measuring the kinetics of GPCR recycling, recent advances in live cell imaging have made detection of GPCR sorting into subdomains of endosomes possible (Puthenveedu et al., 2010; Temkin et al., 2011; Vistein & Puthenveedu, 2013). Recycling of several GPCRs requires a sequence of amino acids in the C-terminal tail of the receptor, and these sequences sort the receptor into actin-stabilized domains at the endosome, distinct from bulk recycling domains at the endosome (Lauffer et al., 2010; Puthenveedu et al., 2010). Recent work shows that signaling kinases can regulate GPCR recycling and resensitization by controlling entry of receptors into specific domains of the early endosome (Vistein & Puthenveedu, 2013). High magnification, live cell confocal fluorescence microscopy allows for visualization of rapid changes in GPCR sorting to these endosomal compartments.
3.3.3.2 Flow of experiment
Plate N-terminally FLAG-tagged receptor, for example, FLAG-B2AR, on #1.5 coverslips. We generate stable cell lines with antibiotic selection, following transfection of FLAG-B2AR with the Effectene transfection reagent, according to the manufacturer’s protocol. If other markers are required, for example, of actin-stabilized endosomal domains, fluorescent protein-tagged versions may be transfected into the FLAG-B2AR stable cell lines. Transfect stable receptor cell lines with actin or other markers 48–72 h prior to performing imaging experiments, and pass cells to coverslips 24–48 h prior to imaging.
Mount coverslips in a live cell imaging chamber. Alternatively, glass bottom dishes can be used.
Replace growth medium (i.e., DMEM + 10% FBS if using HEK 293 cells) with imaging medium (L15 with 5% FBS).
Using confocal fluorescence microscopy with a 100× objective, find a cell that has crisp, even fluorescence of the FLAG-tagged receptor at the cell surface, with very little intracellular receptor, as in Figure 3(A), left image.
To acquire a time course of a large population of endosomes within a cell, capture images every 1 min, and take multiple z sections throughout the cell, with z sections of approximately 0.3 μm through the entire cell. Acquire 1–2 min of baseline surface fluorescence of receptors, then add agonist to activate receptors and induce internalization, and acquire a 15–20 min movie. In the case of FLAG-B2AR, the majority of receptors will be localized to endosomes, as in Figure 3(A), right image.
To image the dynamics of receptor sorting within domains of a single endosome, acquire a fast time course, capturing at least every 1–2 s. Use 0.2 μm sections and capture a total of 1–2 μm of the cell. Because endosomes are mobile, this is necessary to follow endosome events, such as tubule fission and vesicle generation. For example, in Figure 3(C), a FLAG-B2AR tubule persists for approximately 10 s before vesicle fission.
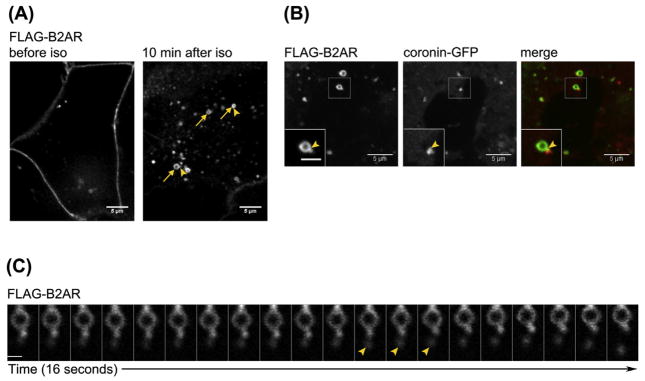
FIGURE 3. Strategy 3: Visualization of sorting and recycling in the endosome.
(A) Example image of FLAG-B2AR localized to the cell surface prior to agonist addition (left panel) and FLAG-B2AR in endosomes 10 min after iso addition (right panel). Endosomes are clearly visible after 10 min of agonist (arrows). Tubule domains are visible protruding from the endosome body (arrowheads). (B) Example image of FLAG-B2AR endosomes imaged with coronin–GFP to mark endosomal actin. A coronin–GFP spot localizes to the base of the FLAG-B2AR tubule, see inset in left corner. Inset scale bar is 2 μm. (C) Example of a FLAG-B2AR tubule fission event imaged every 800 ms. An FLAG-B2AR tubule persists for ~10 s before vesicle fission, and a vesicle is visible next to the endosome (see arrowheads). Scale bar is 1 μm. (See color plate)
3.3.3.3 Considerations
Membrane trafficking events are very temperature sensitive, so it is necessary to image at 37 °C to properly visualize receptor endocytosis and dynamics at the endosome. Also, endosomes move very fast within cells at 37 °C. Therefore, when acquiring z slices in multiple color time-lapse imaging experiments, it is important to minimize delay between channels—i.e., acquire all channels at the same z section before moving to a different plane.
3.3.3.4 Results
Prior to agonist addition, for example, isoproterenol (iso) for B2AR, receptors are primarily localized to the cell surface (Figure 3(A), left panel). Ten minutes after iso addition, the majority of FLAG-B2AR is localized to endosomes (Figure 3(A), right panel). B2AR localizes to tubules characterized by actin spots at the early endosome (Puthenveedu et al., 2010). An actin tubule, marked by coronin–GFP, can be seen at the periphery of the endosome body, at the base of the FLAG-B2AR tubule (Figure 3(B)). Tubule fission events prior to vesicle formation and recycling can be visualized at the early endosome using rapid live cell imaging (Figure 3(C)). In Figure 3(C), a tubule persists for approximately 10 s before fission and vesicle formation occurs.
SUMMARY
In this chapter, we have described live cell imaging protocols for the detection and quantitation of GPCR recycling and endosomal sorting. These strategies can be used to determine kinetic changes in overall rates of GPCR recycling across several cells in response to different agonist and drug treatments. We have also described methods that allow for visualization of individual GPCR recycling events in real time. Additionally, we have explained methods for imaging the dynamics of GPCR sorting at the level of the endosome, as well as how to detect GPCR in different domains of endosomes. Together, these methods are powerful tools that can be used to understand the dynamics and regulation of GPCR trafficking in live cells.
References
- Ambrose EJ. A surface contact microscope for the study of cell movements. Nature. 1956;178(4543):1194. doi: 10.1038/1781194a0. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. Journal of Cell Biology. 1981;89(1):141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SL, Soohoo AL, Shiwarski DJ, Schulz S, Pradhan AA, Puthenveedu MA. Cell-autonomous regulation of muopioid receptor recycling by substance P. Cell Reports. 2015;10(11):1925–1936. doi: 10.1016/j.celrep.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SB, Filardo EJ. Trans-Golgi Network (TGN) as a regulatory node for β1-adrenergic receptor (β1AR) down-modulation and recycling. The Journal of Biological Chemistry. 2012;287(17):14178–14191. doi: 10.1074/jbc.M111.323782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PZC, Gunn P, Gleeson PA. Cargo trafficking between endosomes and the trans-Golgi network. Histochemistry and Cell Biology. 2013;140(3):307–315. doi: 10.1007/s00418-013-1125-6. [DOI] [PubMed] [Google Scholar]
- Drake MT, Shenoy SK, Lefkowitz RJ. Trafficking of G protein-coupled receptors. Circulation Research. 2006;99(6):570–582. doi: 10.1161/01.RES.0000242563.47507.ce. [DOI] [PubMed] [Google Scholar]
- Dunn KW, McGraw TE, Maxfield FR. Iterative fractionation of recycling receptors from lysosomally destined ligands in an early sorting endosome. The Journal of Cell Biology. 1989;109(6 Pt 2):3303–3314. doi: 10.1083/jcb.109.6.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyaloglu AC, McCullagh E, von Zastrow M. Essential role of Hrs in a recycling mechanism mediating functional resensitization of cell signaling. The EMBO Journal. 2005;24(13):2265–2283. doi: 10.1038/sj.emboj.7600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annual Review of Pharmacology and Toxicology. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- Hislop JN, Henry AG, von Zastrow M. Ubiquitination in the first cytoplasmic loop of μ-opioid receptors reveals a hierarchical mechanism of lysosomal down-regulation. The Journal of Biological Chemistry. 2011;286(46):40193–40204. doi: 10.1074/jbc.M111.288555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Zeng X, Kim W, Balasubramani M, Fortian A, Gygi SP, et al. Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(39):15722–15727. doi: 10.1073/pnas.1308014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Wunder C. Retrograde transport: two (or more) roads diverged in an endosomal tree? Traffic. 2011;12(8):956–962. doi: 10.1111/j.1600-0854.2011.01200.x. [DOI] [PubMed] [Google Scholar]
- Lauffer B, Melero C, Temkin P, Lei C, Hong W, Kortemme T, et al. SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. The Journal of Cell Biology. 2010;190:565–574. doi: 10.1083/jcb.201004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, Paing MM, Temple BRS, Trejo J. G protein–coupled receptor sorting to endosomes and lysosomes. Annual Review of Pharmacology and Toxicology. 2008;48(1):601–629. doi: 10.1146/annurev.pharmtox.48.113006.094646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nature Reviews Molecular Cell Biology. 2004;5(2):121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- Mayor S, Presley JF, Maxfield FR. Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. The Journal of Cell Biology. 1993;121(6):1257–1269. doi: 10.1083/jcb.121.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394(6689):192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, et al. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 2010;143(5):761–773. doi: 10.1016/j.cell.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero G, von Zastrow M, Friedman PA. Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: means, motif, and opportunity. Advances in Pharmacology. 2011;62:279–314. doi: 10.1016/B978-0-12-385952-5.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanowitz M, Von Zastrow M. Ubiquitination-independent trafficking of G protein-coupled receptors to lysosomes. The Journal of Biological Chemistry. 2002;277(52):50219–50222. doi: 10.1074/jbc.C200536200. [DOI] [PubMed] [Google Scholar]
- Tanowitz M, von Zastrow M. A novel endocytic recycling signal that distinguishes the membrane trafficking of naturally occurring opioid receptors. The Journal of Biological Chemistry. 2003;278(46):45978–45986. doi: 10.1074/jbc.M304504200. [DOI] [PubMed] [Google Scholar]
- Temkin P, Lauffer, Jäger S, Cimermancic P, Krogan N, Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nature Cell Biology. 2011;13:717–723. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo J. The cytoplasmic tails of protease-activated receptor-1 and substance P receptor specify sorting to lysosomes versus recycling. The Journal of Biological Chemistry. 1999;274(4):2216–2224. doi: 10.1074/jbc.274.4.2216. [DOI] [PubMed] [Google Scholar]
- Vistein R, Puthenveedu M. Reprogramming of G protein-coupled receptor recycling and signaling by a kinase switch. Proceedings of the National Academy of Sciences. 2013;110(38):15289–15294. doi: 10.1073/pnas.1306340110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Dhavan R, Chevalier M, Yudowski G, Zastrow M. Rapid delivery of internalized signaling receptors to the somatodendritic surface by sequence-specific local insertion. The Journal of Neuroscience. 2010;30:11703–11714. doi: 10.1523/JNEUROSCI.6282-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YJ, Arttamangkul S, Evans CJ, Williams JT, von Zastrow M. Neurokinin 1 receptors regulate morphine-induced endocytosis and desensitization of muopioid receptors in CNS neurons. The Journal of Neuroscience. 2009;29(1):222–233. doi: 10.1523/JNEUROSCI.4315-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudowski GA, Puthenveedu MA, Henry AG, von Zastrow M. Cargo-mediated regulation of a rapid Rab4-dependent recycling pathway. Molecular Biology of the Cell. 2009;20(11):2774–2784. doi: 10.1091/mbc.E08-08-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Canals M, Murphy JE, Klingler D, Eriksson EM, Pelayo JC, et al. Agonist-biased trafficking of somatostatin receptor 2A in enteric neurons. The Journal of Biological Chemistry. 2013;288(36):25689–25700. doi: 10.1074/jbc.M113.496414. [DOI] [PMC free article] [PubMed] [Google Scholar]