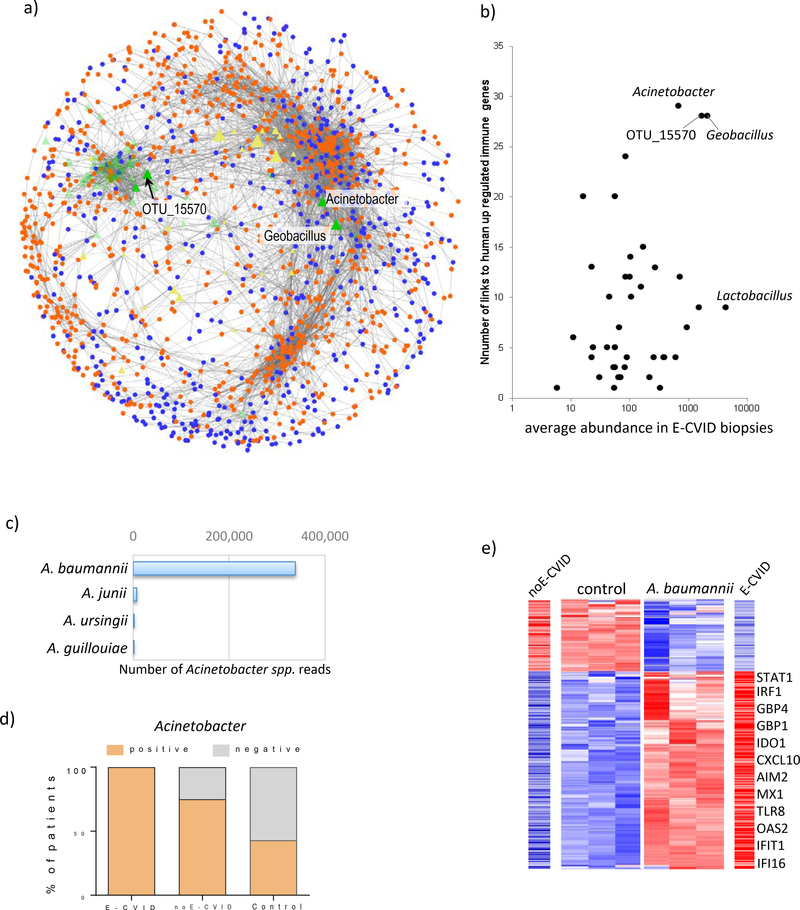
Figure 4.
Transkingdom network analysis identifies Acinetobacter baumannii as a candidate microbe driving CVID enteropathy. (a) Transkingdom network reconstructed from levels of expression of human genes from E-CVID signature and candidate microbes selected based on analysis in Fig. 3d; orange and blue indicate up-regulated and downregulated genes in E-CVID, respectively; green and yellow are bacterial OTUs increased or decreased in E-CVID, respectively. (b) Number of connections to genes up-regulated in E-CVID and average abundance in E-CVID biopsies are plotted for each OTU identified by analysis in Fig. 3d. (c) Number of reads corresponding to species of Acinetobacter genus in biopsies based on shotgun metagenomics. (d) Number of biopsy samples positive or negative for Acinetobacter in each patient group (p<0.04, Chi-square test). (e) Gene expression from E-CVID signature as assessed by RNAseq in THP-1 cells after 6 h of incubation with A. baumannii. Representative genes are indicated and levels of expression of the same genes in biopsies with E-CVID or noE-CVID are shown; full list of genes is in Supplementary Table S13.