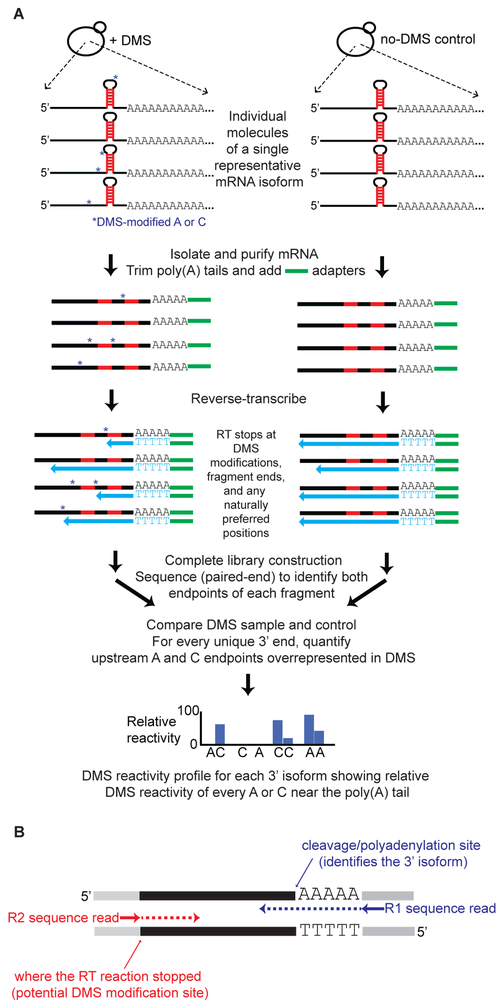
Figure 1. DREADS schematic.
(A) Exponentially-growing yeast cells are treated with DMS, which methylates exposed A and C residues in cellular RNAs. During library construction, reverse transcriptase (RT) stalls at DMS-modified positions (blue asterisks) as well as at naturally occurring stall sites on mRNA molecules. Each resulting library fragment contains information on both the specific poly(A) isoform (downstream end) and the endpoint of the RT reaction (upstream end). Paired-end deep sequencing thus makes it possible to link multiple RT stops with individual 3’ isoforms. Subtraction of the naturally occurring (non-DMS) RT stops in the untreated control allows for precise mapping of DMS-reactive, accessible A and C residues in each 3’ isoform.
(B) Example of a library fragment subjected to paired-end sequencing. Gray segments at the fragment ends represent adapter sequences added during library construction. The R1 sequence read encompasses the junction between the poly(A) tail remnant and the last nt of genomically-encoded 3’UTR sequence; this allows mapping of the specific 3’ isoform. The R2 sequence read supplies information about where the reverse transcription reaction stopped (naturally or at potential DMS-modified site) on the same mRNA molecule during library construction.