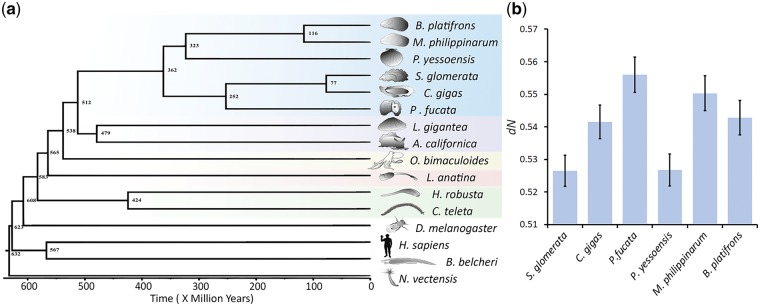
Figure 1.
Divergence time and rate of non-synonymous substitutions between bivalves. (a) A time tree based on protein sequences from 16 metazoan genomes. Divergence times were estimated using a Bayesian MCMC method. A maximum likelihood based phylogenetic tree was calibrated based on seven well-defined fossils (see Methods). (b) Genetic divergence measured by the non-synonymous substitution rate (dN) between bivalves and the outgroup Lottia gigantea using 3,269 orthologous genes. The relative divergences with respect to the outgroup reveal the differences in the rate of protein evolution among bivalves.