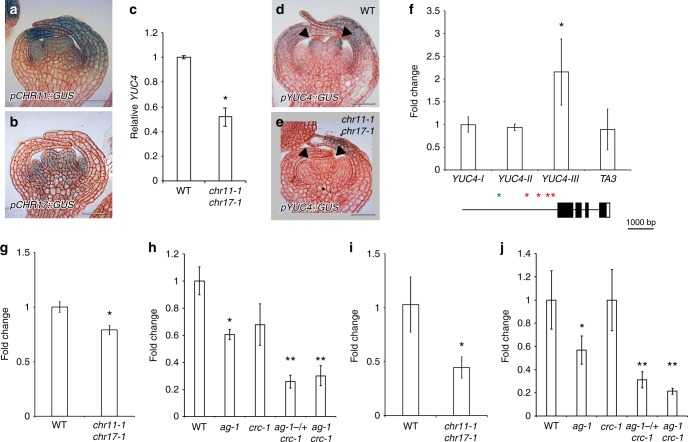
Fig. 4.
AG and CRC are required for increased chromatin accessibility at the YUC4 loci. a, b Expression of pCHR11::GUS (a) and pCHR17::GUS (b) in stage 6 floral buds. c YUC4 mRNA levels in wild-type and chr11-1 chr17-1 floral buds. The values are represented as the means ± SEMs. p-values were calculated with a two-tailed Student’s t-test. *p < 0.05 compared to the wild type. d, e Expression of pYUC4::GUS in wild-type (d) and chr11-1 chr17-1 (e) floral buds at the stage 6. Arrowheads indicate abaxial side of epidermal cells in carpels at stage 6. f Top: Mapping of the GFP-CHR11 protein association to the YUC4 locus. A GFP antibody was used to detect CHR occupancy. Bottom: Diagram of ChIP-qPCR amplicons tested. Red and green asterisks indicate CRC binding sites and possible CArG box (C[C/T][A/T]G[A/G][A/T]6[A/G]G) within an AG binding peak, respectively. The values are represented as the means ± SEMs. p-values were calculated with a Student’s t-test. *: p < 0.05 compared to the TA3 signal. g, h Chromatin accessibility at the YUC4 locus. g) Wild-type and chr11-1 chr17-1 floral buds. h) Wild-type, ag-1, crc-1, ag-1-/ + crc-1, and ag-1 crc-1 floral buds. The values are represented as the means ± SEMs. p-values were calculated with a two-tailed Student’s t-test. *p < 0.05 compared to the wild type. **p < 0.05 compared to single mutants. i, j RNA Pol II association with the YUC4 locus. A Pol II antibody was used. i Wild-type and chr11-1 chr17-1 floral buds. j Wild-type, ag-1, crc-1, ag-1-/+ crc-1, and ag-1 crc-1 floral buds. The values are represented as the means ± SEMs. p-values were calculated with a two-tailed Student’s t-test. *p < 0.05 compared to the wild type. **p < 0.05 compared to single mutants. Bars = 50 µm in a, b, d, e