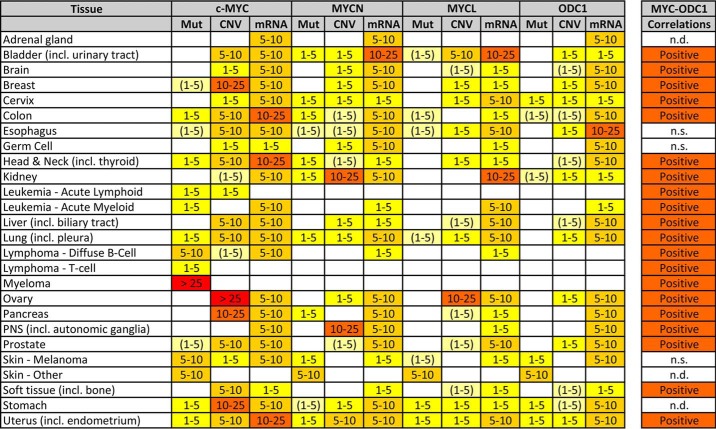
Table 1.
MYC and ODC1 aberrations in human cancer
Public human cancer data were queried for coding mutations (Mut), copy number variations (CNV), and mRNA dysregulation (mRNA) of the (three) MYC and ODC1 genes. Numbers represent % of samples with an aberration: white fields represent <1% aberrations; colored fields represent 1–5, 5–10, 10–25, and >25%. (1–5) means that 1–5% aberrations were found in specific tumor subtypes only. All datasets containing human cancer samples available on the public websites COSMIC (https://cancer.sanger.ac.uk/cosmic), cBioPortal (http://www.cbioportal.org/) (167, 168), and OncoMine (https://www.oncomine.org/) (169) were analyzed for a total of >1000 sets. Tumor types are represented by a minimum of three datasets. (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.) Aberrations from identical samples in different datasets were not over-counted. Single mutations in a dataset or datasets containing <100 samples, were not used for calculations. Presented are only the significant changes in mutant reads, CNV, or mRNA expression, according to the website default analysis parameters. Correlations between (any) MYC and ODC1 mRNA expressions were calculated on the R2 website (http://r2.amc.nl/), using a 2log Pearson test on all tumor datasets on Affymetrix arrays (n = 242). Analysis was as in Ref. 17. n.d. = no data, n.s. = not significant.