FIGURE 9.
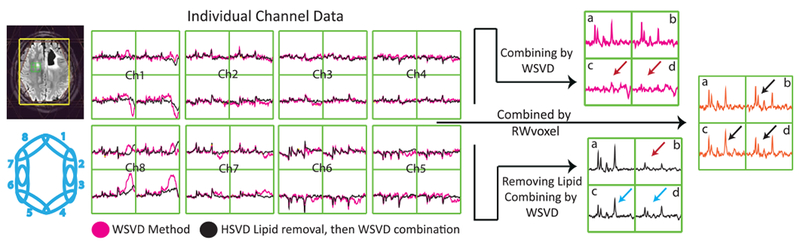
Impact of lipid contamination on WSVD combination. Left: the individual channel data are shown before (in magenta) and after (in black) HSVD lipid removal. Large lipid signals are observed in channels 1and 8 of the uncombined spectra that will impact the overall combination on the right. The HSVD lipid removal algorithm clearly failed for the top right voxel in channels 1 and 8, such that it ends up with higher lipid levels than are present in the original data. Right: the combined spectra using the WSVD alone and WSVD in addition to lipid removal. When combining the spectra using WSVD alone (magenta), voxels a and b have good SNR but voxels c and d lack any signal. Incorporating lipid removal (black spectra) improves voxels c and d at the expense of voxel b
