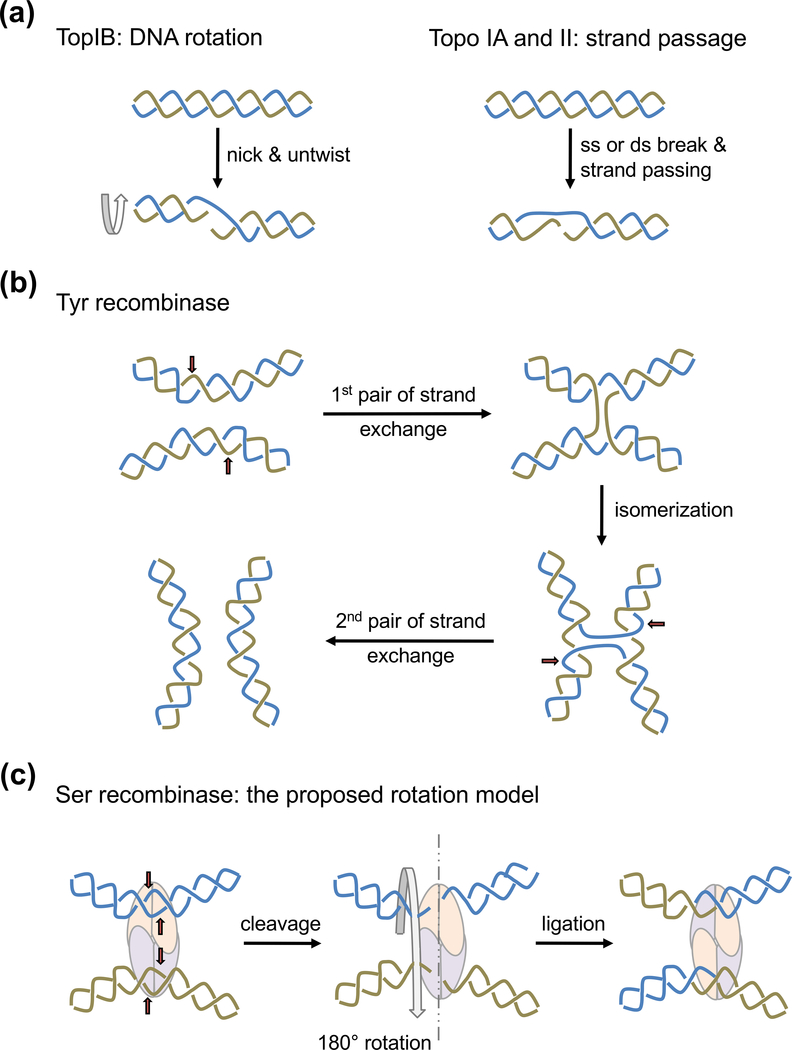
Fig. 2.
Mechanisms of topoisomerization and recombination by YRs. (a) Two mechanisms are used for topoisomerization. Type IB topoisomerases (TopIB) relax (−) or (+) supercoils by nicking one strand and allowing DNA to untwist. Topo IA and Topo II can relax as well as make supercoils by breaking one strand or duplex and passing the other through it. The intertwined blue and beige color lines represent single strands in the case of Topo IA or double helices in the case of Topo II. (b) Tyrosine recombinases (YRs) catalyze DNA recombination in two steps. One pair of strands are cleaved, exchanged and rejoined in each step (beige and blue colored lines sequentially). The red arrows point the cleavage sites. A Holliday junction is formed and isomerized between the two steps. (c) Serine recombinases make concerted double strand breaks of two recombination sites. The proposed subunit rotation mechanism requires the left half of the cleaved DNA rotate 180° relative to the right half for strand exchange.