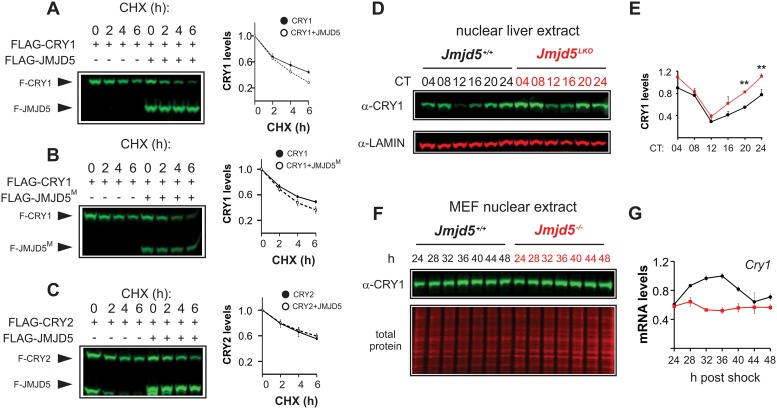
Fig 3. JMJD5 destabilizes CRY1.
Impact of (A) wild-type and (B) catalytically inactive JMJD5M on CRY1 stability. (C) JMJD5 is unable to degrade FLAG-CRY2. Densitometric analysis of independent experiments for A–C are shown next to the corresponding blots (mean ± SEM, n = 4, n = 3, and n = 3 respectively). Please note the presence of an unknown lower molecular weight band observed in FLAG-CRY2 transfections. (D) JMJD5-null livers have increased levels of endogenous nuclear CRY1 protein across the circadian cycle. (E) Densitometric analysis of CRY1 levels in control and JMJD5-null livers corresponding to (D) (mean ± SEM, n = 3). (F) CRY1 levels in nuclear extracts of Jmjd5+/+ and Jmjd5−/− MEFs. Cry1 mRNA levels from Fig 1 have been replotted to span 24–48 hours post synchronization. CHX, cycloheximide; CRY, CRYPTOCHROME; JMDJ5, JmjC domain–containing protein 5; Jmjd5LKO, Jmjd5 liver knockout; JMJD5M, JMJD5 mutant; MEF, mouse embryo fibroblast.