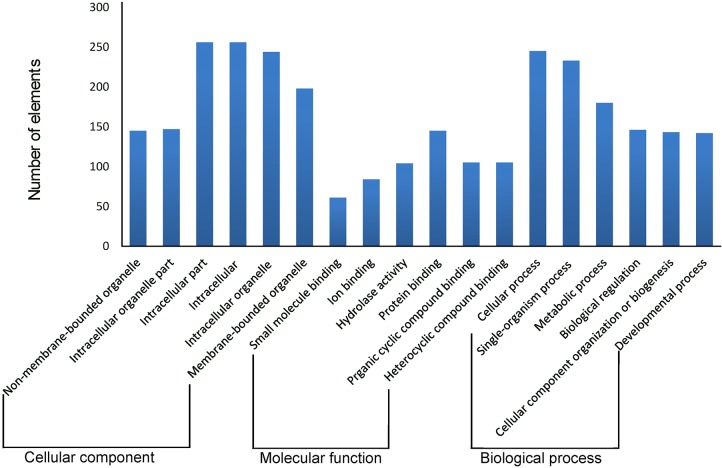
Fig 4. Gene ontology classification of G. duodenalis genes.
Distribution of the GO categories assigned to the G. duodenalis genomes. Genes were classified into three categories: cellular components (Level 3), molecular functions (Level 3) and biological processes (Level 2). Regarding the GO annotation, 921 (50%) SSR loci were validated by the Blast2GO annotation procedure, resulting in 2,793 GO terms: cellular component terms composed most of the functional vocabulary (33.90%), followed by the molecular function (33.62%) and biological process (32.47%) classes. Within the biological process category, metabolic processes (99), protein phosphorylation (87) and the serine family amino acid metabolic process (71) figured prominently. Regarding the molecular function category, ATP binding (181) and protein serine/threonine kinase activity (66) were the most represented GO terms. Half of the cellular component class of annotations was related to integral components of the membrane (465).