Abstract
Remodeling of membranes by fission or fusion has been extensively studied in eukaryotes, but proteins directly responsible for mediating such events in bacteria have not been discovered. A recent report identified a protein in Bacillus subtilis that exploits an affinity for a specific lipid to drive membrane fission during sporulation.
Membrane remodeling is an integral part of numerous biological processes found in all domains of life. Remodeling of membranes occurs largely through two processes: membrane fission where one membrane divides into two and membrane fusion where two membranes come together to form one. The discovery of SNARE proteins that facilitate membrane fusion [1] and the dynamin protein family [2] and endosomal sorting complex for transport (ESCRT-III complex) [3] which facilitate membrane fission have led to a better understanding of the mechanisms that govern membrane fusion and fission in eukaryotes (depicted in Fig. 1). However, there are still many factors that regulate and participate in membrane remodeling that remain elusive. Discriminating between factors that are directly responsible for membrane remodeling and factors that are necessary for the events that precede or follow the membrane remodeling event has been a challenge due to the formation of interdependent complexes at points of membrane fission and fusion. In particular, the specific factors involved in prokaryotic membrane remodeling remain a mystery largely because the factors that may mediate these processes are likely essential for viability. For example, despite the identification and characterization of many factors required for prokaryotic cell division, the factors directly responsible for membrane fission in this process are unknown. A new study by Doan et al. [4] has discovered the first protein that has been shown to directly mediate membrane remodeling during spore formation in the bacterium Bacillus subtilis.
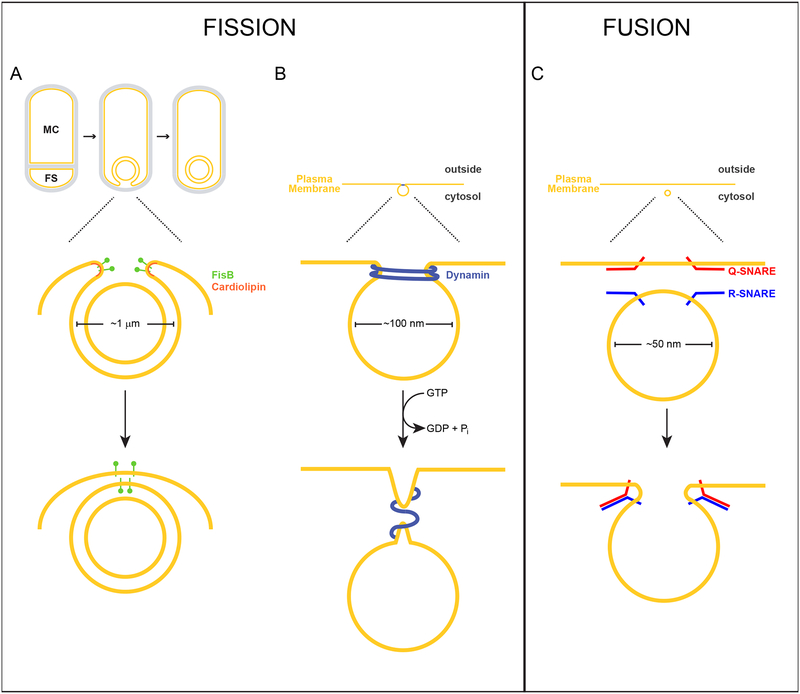
Figure 1. Membrane fission of the engulfing sporulation membrane mediated by FisB, or endocytic vesicles mediated by dynamin, and membrane fusion of synaptic vesicles mediated by SNARE complexes.
Above, scale drawings of a 1 μm diameter developing forespore of the bacterium Bacillus subtilis, 100 nm diameter endocytic vesicle, and a 50 nm diameter synaptic vesicle are depicted. Lipid bilayers are depicted in yellow; peptidoglycan is depicted in gray. (A) Sporulation in B. subtilis initiates with an asymmetric division event that divides the cell into two genetically identical compartments: the mother cell (MC) and forespore (FS). Next, the asymmetric septum curves as the mother cell engulfs the forespore. Eventually, the leading edges of the engulfing membrane undergo a membrane fission event, pinching off the forespore as a double membrane-bound organelle in the mother cell. FisB (green) localizes to the site of fission, where its extracellular domain interacts with the lipid cardiolipin (orange) which may be enriched along the concave (negatively curved) inner leaflet of the phospholipid bilayer at this site. (B) Dynamin (blue) assembles at the neck of an endocytic vesicle as it forms from the plasma membrane. GTP hydrolysis drives the constriction of dynamin, driving membrane scission. (C) During exocytosis, R-SNAREs anchored in the vesicular membrane interact with Q-SNARES on the target membrane to form a stable cis-SNARE complex that drives membrane fusion.
When B. subtilis sense nutrient deprivation the cells undergo a simple developmental program called sporulation, which results in the production of a largely dormant cell type that protects the cell’s genetic material until favorable growth conditions are restored [5, 6]. The rod-shaped bacterium first differentiates into two genetically identical, but morphologically distinct cells through an asymmetric septation event which results in a larger cell (the “mother cell”) and a smaller cell (the “forespore”) that lie side by side (Fig. 1, top center). Next, the mother cell begins to engulf the forespore in a phagocytic-like event. At the end of engulfment the leading edges of the engulfing membrane undergo a membrane fission event that pinches off the forespore as a free floating double-membrane bound organelle inside the mother cell cytosol. The mother cell nourishes the forespore as it matures and eventually undergoes a programmed lysis event that results in the spore being released into the environment. Factors involved in sporulation are often non-essential for normal growth, thus facilitating the identification of novel factors whose homologs may participate in other essential biological processes [7]. In their study, Doan et al. sought to identify factors that catalyze membrane fission in prokaryotes by focusing on the sporulation-specific fission event that occurs at the end of engulfment.
After ruling out likely factors that could catalyze the membrane fission step of engulfment, the authors employed a candidate approach in which they hypothesized that the gene encoding the fission factor must satisfy three requirements: 1) it would need to encode a transmembrane protein, 2) it would be expressed in the mother cell before the onset of engulfment, and 3) deletion of the gene would result in a sporulation defect. Using a fluorescence microscopy-based screen of a library of deletion mutants that fit their criteria, the authors identified a mutant strain which was significantly delayed in the completion of engulfment. The previously uncharacterized gene was renamed fisB (fission protein B). Cells lacking FisB had wild type rates of membrane migration during engulfment, but were unable to complete engulfment presumably due to a loss in the ability to undergo membrane fission. Interestingly, the authors noted that while most ΔfisB cells were defective in membrane fission, there was a subpopulation that was delayed in the sporulation program and was able to eventually complete engulfment, suggesting that there may be an alternative fission mechanism. This sort of functional redundancy has also been observed in other steps of sporulation [8] and may be the result of the selection for a robust developmental program to ensure cell survival.
To confirm that FisB is responsible for facilitating membrane fission, the authors constructed a functional GFP-FisB fusion and checked if FisB localizes to the right place at the right time to do its proposed job. When expressed under its native promoter and ribosome-binding site GFP-FisB localized as discrete foci in all membranes, but was enriched in the forespore membrane. To further pinpoint FisB’s subcellular location, the authors expressed a YFP-FisB fusion at lower levels to reduce nonspecific and/or low-affinity binding. As cells neared completion of engulfment, YFP-FisB localized as discrete foci at the mother cell distal side of the forespore - the expected site of the membrane scission event.
To determine if FisB is sufficient for membrane remodeling, the authors employed a classical in vitro assay designed to measure membrane fusion between two liposomes, in which fluorescence quenching is relieved when an unlabeled liposome fuses with a liposome that is loaded with a fluorescent lipid and its quencher [9]. Purified FisB, when incorporated into liposomes, was indeed capable of carrying out lipid mixing in this assay, suggesting that it is sufficient for membrane remodeling. Interestingly, while FisB was able to catalyze lipid mixing in liposomes containing a mixture of lipids that mimicked that of sporulating B.subtilis, it was unable to do so in liposomes composed of a minimal eukaryotic lipid mix routinely used for SNARE-mediated fusion assays. By using a coflotation assay, the authors determined that FisB preferentially interacted with cardiolipin, a lipid species that preferentially partitions to highly negatively curved membranes in bacteria and is coincidentally enriched at the forespore during sporulation in B. subtilis. Unlike previously identified cardiolipin interacting proteins, FisB did not interact with other negatively charged lipids [10] suggesting a novel mechanism of protein-cardiolipin interaction. Additionally, the authors mixed labeled FisB liposomes with FisB containing or protein-free liposomes and discovered membrane fusion only required FisB to be present in one lipid membrane. Taken together, the authors proposed a model in which FisB and cardiolipin interactions in the leading edge of the engulfing membrane cause a destabilization of the membrane, which then leads to scission of the engulfing membrane.
Doan et al. have thus described the discovery and characterization of the first protein shown to directly function in prokaryotic membrane remodeling. FisB appears to facilitate membrane scission in a distinct mechanism that involves lipid-specific interactions that can occur in trans. It will be interesting to determine if there are proteins participating in other prokaryotic membrane remodeling events that function through a mechanism similar to FisB. As with many developmental processes in biology that rely on multiple and/or redundant mechanisms, membrane fission cannot be completely eliminated by removing specific factors, as seen by the ability of the cell to complete engulfment even in the absence of FisB or reduced levels of cardiolipin. Mitochondria, which are thought to have originated from endosymbiotic bacteria, are constantly undergoing fusion and fission to maintain homeostasis [11]. It is known that the dynamin-related proteins (Drp1 in humans and Dnm1 in yeast) are responsible for generating the constricting force for mitochondrial fission, but other associated proteins are also necessary [12]; perhaps there are as of yet undiscovered redundant proteins that participate in mitochondrial membrane fission that can exploit a FisB-like affinity for cardiolipin. Additionally, recent studies on L-form bacteria that lack peptidoglycan and the bacterial tubulin homolog FtsZ, which are both essential for cell division in non-L-form bacteria, have been observed to successfully undergo membrane fission [13] further suggesting that there are still undiscovered pathways involved in prokaryotic membrane remodeling.
Acknowledgments
Funding provided by the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research
REFERENCES
- 1.Sudhof TC, and Rothman JE (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campelo F, and Malhotra V (2012). Membrane fission: the biogenesis of transport carriers. Annual review of biochemistry 81, 407–427. [DOI] [PubMed] [Google Scholar]
- 3.Hurley JH, Boura E, Carlson LA, and Rozycki B (2010). Membrane budding. Cell 143, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doan T, Coleman J, Marquis KA, Meeske AJ, Burton BM, Karatekin E, and Rudner DZ (2013). FisB mediates membrane fission during sporulation in Bacillus subtilis. Genes & development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stragier P, and Losick R (1996). Molecular genetics of sporulation in Bacillus subtilis. Annual review of genetics 30, 297–241. [DOI] [PubMed] [Google Scholar]
- 6.Errington J (2003). Regulation of endospore formation in Bacillus subtilis. Nature reviews 1, 117–126. [DOI] [PubMed] [Google Scholar]
- 7.Fay A, and Dworkin J (2009). Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. Journal of bacteriology 191, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broder DH, and Pogliano K (2006). Forespore engulfment mediated by a ratchet-like mechanism. Cell 126, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, and Rothman JE (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- 10.Mileykovskaya E, and Dowhan W (2009). Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochimica et biophysica acta 1788, 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youle RJ, and van der Bliek AM (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lackner LL, Horner JS, and Nunnari J (2009). Mechanistic analysis of a dynamin effector. Science 325, 874–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, and Errington J (2009). Life without a wall or division machine in Bacillus subtilis. Nature 457, 849–853. [DOI] [PubMed] [Google Scholar]