Abstract
Oncogenic lesions are surprisingly common in morphologically and functionally normal human skin, however, the cellular and molecular mechanisms that suppress their cancer-driving potential to maintain tissue homeostasis are unknown. By employing assays for direct and quantitative assessment of cell fate choices in vivo, we show that oncogenic activation of PI3K/AKT, the most commonly activated oncogenic pathway in cancer, promotes differentiation and cell-cycle exit of epidermal progenitors. As a result, oncogenic PI3K/AKT activated epidermis exhibits growth disadvantage even though its cells are more proliferative. To uncover the underlying mechanism behind oncogene-induced differentiation, we conduct a series of genetic screens in vivo, and identify an AKT substrate SH3RF1 as a specific promoter of epidermal differentiation that has no effect on proliferation. Our study provides evidence for a direct, cell autonomous mechanism that can suppresses progenitor cell renewal and block clonal expansion of epidermal cells bearing a common and activating mutation in Pik3ca.
Uncontrolled tissue expansion is the fundamental feature of cancer thought to be acquired early in tumorigenesis1. Oncogenes are thought to be the major drivers of growth in cancer. They fuel tissue expansion by accelerating cell cycle, inhibiting cell death or cell cycle arrest, and are also believed to promote self-renewal of tissue stem cells1,2. Yet, skin epidermis, the tissue of origin for basal and squamous cell carcinomas, was recently shown to contain an abundance of cells carrying activated oncogenes without any consequences. Moreover, clones containing activated oncogenes did not appear to be under positive selection, and clonal expansion was only associated with lesions in a small set of genes3. This surprising observation indicated that tumor suppressive mechanisms are at work early on in epidermis to counteract growth-promoting functions of activated oncogenes and keep clonal expansion under stringent control.
Senescence is a well-documented growth suppressive mechanism by which cells respond to an oncogenic stress in a cell autonomous manner. Activation of BRAF V600E in nevi4 and NRAS in lymphocytes5 can induce cellular senescence in vivo. Similarly, loss of the tumor suppressor PTEN, which activates the oncogenic PI3K/AKT signaling, induces senescence in prostate epithelium6. Apoptosis can also suppress oncogene driven clonal expansion, as demonstrated following overexpression of MYC and E1A in cell culture7–9. While highly efficient at restricting oncogene-driven growth, both senescence and apoptosis bring on a dramatic loss of cells and their growth potential. As such, they are not compatible with observations that skin epithelium maintains its structure, function, and rapid turnover despite the massive amount of oncogenic lesions.
Growth in skin epithelium is driven by progenitor cells that can self-renew, to maintain tissue’s growth potential, and differentiate into postmitotic progeny, which provide the form and function of skin10,11. Although these cell fate decisions directly control the number of progenitors in a tissue over time, and are therefore a critical determinant of its growth potential1,12, whether they contribute to regulation of clonal expansion in the context of oncogenic stress is not known.
The PI3K/AKT pathway is commonly hyper-activated in cancers13, and suppression of PI3K signaling has been shown to significantly inhibit proliferation and cell survival in epidermal squamous cell carcinoma (SCC)14,15. Yet, despite the observation that oncogenic mutations in the PI3K/AKT pathway are among the most common lesions in SCCs and robust PI3K/AKT activity is also detected in premalignant epithelia16,17, very little is known about how they affect clonal expansion. In addition, the effect of normal PI3K/AKT signaling on epidermal stem cell renewal seems to be context-dependent. Activation of PI3K in organotypic culture of epidermal progenitors was shown to promote colony formation and decrease differentiation marker expression18,19, whereas in conventional culture condition, it had the opposite effect20,21. In epidermal development, inhibition of PI3K/AKT signaling was shown to suppress expression of the progenitor cell marker TP6322, while loss of PDK1, the upstream activator kinase of AKT, resulted in increased TP63 expression23. Despite the different effect on TP63 expression, both studies reported that suppression of PI3K/AKT signaling blocked epidermal stratification. In the adult, expression of myrAkt was shown to promote expansion of hair follicle stem cells24,25, supporting the longstanding idea that oncogenes drive stem cell renewal to contribute to tumorigenesis2. Importantly, the effect of oncogenic mutations in the PI3K/AKT pathway on epithelial progenitor cell renewal and differentiation in adult epidermis, where tumors usually originate, has never been directly tested.
In the current study, we show that oncogene induced-differentiation is the dominant growth suppressive mechanism in oncogenic Pik3ca-activated epidermis that restricts clonal expansion. Using independent and direct measurements of cell fate choice in both fixed tissues and live animals, we show that oncogenic activation of PI3K in adult epidermis results in a cell autonomous suppression of symmetric renewal that drives reduced clonal expansion and long-term loss of oncogene-expressing epithelial cells. We employ a series of genetic screens in vivo, and uncover an AKT substrate SH3RF1 as a specific promoter of progenitor renewal that has no effect on proliferation. Lastly, we use tests of genetic epistasis in vivo to show that oncogenic activation of PI3K signaling results in AKT-mediated suppression of SH3RF1 scaffold function in supporting pro-renewal JNK signaling.
Results
Oncogenic activation in PI3K pathway inhibits clonal expansion.
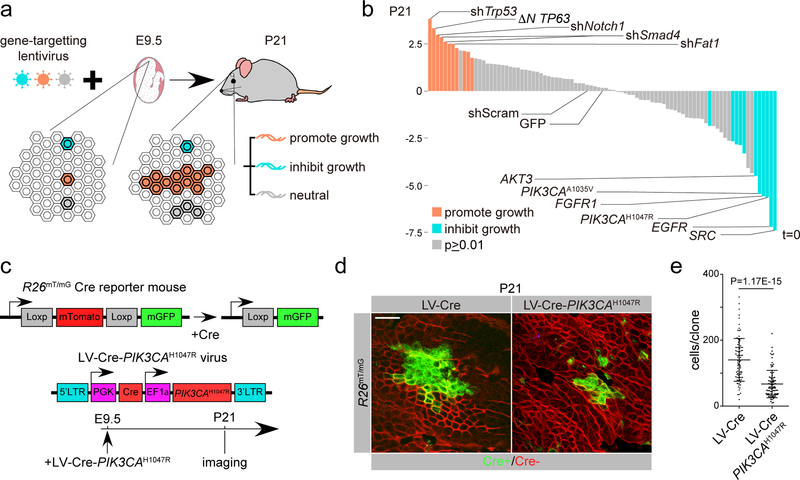
We selected 35 known drivers of squamous cell carcinomas3 (SCCs; Supplementary Fig. 1a-c), and generated ORF- and shRNA-expressing constructs to model their gain- and loss-of-function lesions. To test how these cancer drivers affect epithelial growth, we introduced them as a lentiviral pool into mouse epidermis via ultrasound-guided in utero microinjection26,27. We reasoned that constructs that impact growth would become enriched or depleted in the epidermis over time, which we could measure by sequencing27 (Fig. 1a). Consistent with previous findings, lesions associated with clonal expansion in human skin3 were among our top promoters of epidermal growth (Fig. 1b; Supplementary Table 1). Unexpectedly, our screen also identified several oncogenes in the PI3K/AKT pathway as significant suppressors of growth (Fig. 1b). We focused on Pik3ca, which encodes the catalytic subunit of PI3K and is the most commonly mutated proto-oncogene across epithelial cancers13,28,29 (Supplementary Fig. 1d). To independently measure the impact of oncogenic Pik3ca on epithelial growth we transduced epidermis of Cre-reporter mice (R26mT/mG;30) with lentivirus expressing Cre-recombinase (LV-Cre) alone or together with oncogenic Pik3ca (Pik3caH1047R) at clonal density. We observed that oncogenic Pik3ca suppressed epidermal growth and clonal expansion, thus validating our genetic screen (Fig. 1c-e).
Fig. 1. Oncogenic activation of the catalytic subunit of PI3K inhibits clonal expansion.
a, Schematic of a genetic screen for regulators of clonal expansion, based on enrichment (orange) or depletion (blue) of individual barcoded gene-targeting lentivirus over time. b, Needle plot shows enrichment of growth promoting (orange) and growth inhibiting (blue) lentivirus in P21 epidermis relative to their initial abundance in the lentivirus pool (t=0). shScram and GFP are control constructs unchanged between t=0 and P21. Fold change shown is an average value from 3 biological replicates. c, Cre reporter mouse exchanges membrane-associated red (mT) fluorophore with green (mG) following Cre expression. Cre recombinase is expressed from a lentivirus that co-expresses PIK3CAH1047R ORF. d, Representative image of wild-type (LV-Cre) and oncogenic Pik3ca (LV-Cre-PIK3CAH1047R) clones in P21 epidermis of Cre-reporter (R26mT/mG) mice. Untransduced epidermis (red) surrounds transduced (green) cell clone. Scale bar, 100 μm. e, Quantification of cell numbers per clone in P21 epidermis demonstrates that oncogenic PI3K activation results in significant reduction in clone size relative to wild-type. Statistics were derived from n=91 WT and 90 PIK3CAH1047R clones pooled from 3 animals of each condition. Two-tailed t test, P-value<0.0001. Error bar: SD, center value: mean. Statistical source data for e are shown in Supplementary Table 4.
PI3K activation promotes differentiation of epidermal progenitor cells.
To probe how oncogenic Pik3ca inhibits clonal expansion, we turned to a conditional knock-in mouse31–33 for expression of activated Pik3ca at physiological levels and under endogenous control (Fig. 2a). We established that Cre-recombinase efficiently replaces the wild-type protein with its oncogenic form, and results in activation of AKT in Pik3caH1047R/H1047R (from hereon Pik3ca 2X) epidermis (Supplementary Fig. 2a,b). We next measured progenitor cell proliferation, and observed a significant ~2-fold increase in Pik3ca 2X tissues compared to wild-type during perinatal and adult stages (Fig. 2b and Supplementary Fig. 2c,d). We also tested for apoptosis and senescence but detected no change in either (Fig. 2c,d). Increased proliferation not balanced by apoptosis or senescence stood in contrast to our observation that oncogenic Pik3ca inhibited clonal growth, and suggested additional mechanisms of growth inhibition.
Fig. 2. Oncogenic PI3K activation promotes epidermal differentiation but not apoptosis or senescence.
a, Schematic of Pik3caH1047R/H1047R knock-in mouse. b, Quantifications of the rate of cell division, measured by EdU incorporation and average division interval (see Supplementary Fig. 2d and Methods), show that basal progenitors in Pik3ca 2X epidermis divide faster than wild-type (WT) at P21. Statistics based on n=3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. c, Quantification of activated Caspase3 staining shows no significant difference between Pik3ca 2X and WT epidermis at P21. Statistics based on n=3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. d, Senescence associated β-gal (SA-βgal) staining and Cdkn2a expression failed to detect any senescent cells in WT and Pik3ca 2X epidermis at P21. Statistics based on n=3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. ND: not detected. e, Schematic of the EdU-BrdU pulse-chase differentiation assay. EdU only progeny cells (red), labeled by the initial two-hour pulse of EdU, are assessed for expression (brown) or absence (blue) of differentiation marker K10, six hours after completing the S-phase. Rate of renewal is expressed as the fraction of new K10 negative progenitors out of the total progeny. f, Oncogenic Pik3ca promotes differentiation in epidermis at embryonic, perinatal, and adult stages. Statistics based on n=3 animals of each condition. Error bar: SD, center value: mean. g-i GSEA (g) shows that oncogenic Pik3ca biases basal progenitor cell transcriptome towards differentiation. ES, NES, FDR generated by GSEA2 program. Gene ontology enrichment (h) in genes up- and down-regulated in Pik3ca 2X basal cells. Enrichment P-value generated by PANTHER. Expression of self-renewal and differentiation markers in WT and Pik3ca 2X basal progenitors (i). Statistics are based on RNA-seq performed in n=3 animals of each condition. g-i represent data pooled from 3 RNA-seq replicates. Statistical source data for b-d,f are shown in Supplementary Table 4.
Proliferation in wild-type and Pik3ca 2X epidermis was restricted to Keratin 10 (K10) negative basal progenitors (Supplementary Fig. 3a,b). We reasoned that an increase in their differentiation into post-mitotic progeny would contract the tissue’s proliferative compartment, and therefore suppress growth. To test if oncogenic Pik3ca affects differentiation, we developed an assay of progenitor renewal based on EdU-BrdU pulse-chase incorporation (Fig. 2e). We established that a two-hour EdU pulse followed by a six-hour BrdU chase allowed for >95% of EdU+-only progenitors to complete mitosis (~3% remain in G2), and show their daughter cell fate based on expression of differentiation marker K10 in both wild-type and Pik3ca 2X epidermis (Supplementary Fig. 3c,d). We readily detected EdU+-only cells and their differentiation status, and used the fraction of K10- progenitors in the EdU+-only daughter cell population as a measure of progenitor cell renewal rate in vivo (Fig. 2e, Supplementary Fig. 3e).
Using EdU-BrdU pulse-chase assay, we first measured progenitor renewal rate in wild-type epidermis, and found that it ranges from ~0.75 at birth to ~0.5 in the adult (P21-P84, Fig. 2f). Strikingly, we observed a significant decrease in progenitor renewal in Pik3ca 2X epidermis across all stages (Fig. 2f), suggesting that expression of oncogenic Pik3ca promotes differentiation. Importantly, increased differentiation was independent of cell crowding (Supplementary Fig. 4a) or if oncogenic Pik3ca was expressed clonally or throughout the epithelium, suggesting a cell autonomous mechanism (Supplementary Fig. 4b,c). In addition, differentiation marker expression and tissue turnover rates were similar in control and Pik3ca 2X epidermis (Supplementary Fig. 4d,e), indicating that oncogenic Pik3ca promotes differentiation by regulating the initial cell fate choice. Interestingly, we found that renewal rate of wild-type progenitors adjacent to Pik3ca 2X clones (≤5 cells from Pik3ca 2X clone) was elevated (Supplementary Fig. 4b,c). This was not seen in cells that are nearby but non-adjacent to Pik3ca 2X clones, and suggested that local increase in renewal of wild-type progenitors could balance the loss of oncogene-expressing progenitors (Supplementary Fig. 4a-c).
Next, we performed RNAseq on FACS-isolated α6-Integrin expressing (α6 Itghigh) basal progenitors from wild-type and Pik3ca 2X epidermis and found that oncogenic Pik3ca shifts the progenitor expression profile towards differentiation in both mouse34,35 and human36 (Fig. 2g and Supplementary Fig. 4f). Gene ontology (GO) analysis further revealed increased expression of cell cycle regulators, and down-regulation of negative regulators of differentiation in basal progenitors in Pik3ca 2X epidermis (Fig. 2h). Increased differentiation in Pik3ca 2X progenitors was also evidenced by expression of differentiation markers and regulators, including suppression of Id237, Sox938 and elevation of Notch1,339 and KRT1,1040 (Fig. 2i). Together, our data demonstrate that oncogenic Pik3ca promotes increased differentiation and cell cycle exit of progenitor cells in the epidermis.
PI3K activation directly modifies cell fate choice in the epidermis to inhibit renewal.
In epidermis, cell fate choice is associated with progenitor cell divisions which give rise to two mitotically active daughters (symmetric renewal), two post-mitotic daughters (symmetric differentiation), or divide asymmetrically to maintain one progenitor cell and generate one differentiated daughter. To probe if oncogenic Pik3ca promotes differentiation by regulating progenitor cell fate choice, we used two-photon intravital microscopy (Supplementary Fig. 5a). To visualize dividing progenitors and follow the fate of their progeny over time, we crossed mice with an epidermal-specific photo-activatable H2B-mCherry41 transgene (K14-H2B-PAmCherry) to Pik3ca 2X mice, before we broadly activated oncogene expression using high-titer transduction with LV-Cre co-expressing membrane-associated GFP (LV-Cre-mGFP; Fig. 3a and Supplementary Fig. 5b). At P21, we photo-labeled individual GFP+ progenitors (Fig. 3b,c), and followed them as they divided (division 1). We tracked their daughter cells, which we scored as progenitors if they maintained mitotic potential, evidenced by cell cycle re-entry (division 2), or differentiated progeny if they left the basal layer and assumed flattened morphology (Fig. 3b,c). We observed that epidermal progenitors in wild-type skin undergo all three types of cell divisions, with asymmetric being more common (~50%) followed by similar rates of symmetric renewal (~25%) and differentiation (~25%, Fig. 3d). In contrast, symmetric renewal was significantly suppressed in oncogenic Pik3ca-expressing epidermis, whereas asymmetric division and symmetric differentiation were increased (Fig. 3d).
Fig. 3. Oncogenic PI3K/AKT induces differentiation via suppression of progenitor self-renewal.
a, Mouse for epidermis-specific expression of photoactivatable mCherry transgene is used in conjunction with a lentivirus for expression of membrane-associated GFP and Cre recombinase. b, Schematic of an intravital imaging experiment to label individual epidermal progenitors by photo-activating H2B-PAmCherry, and follow them as they divide, and their daughter cells commit to renewing or differentiating fate choices. c, Membrane-associated GFP (green) that marks the epidermal field is co-expressed with Cre-recombinase in a lentivirus (LV-Cre-mGFP). The progeny of the photo-activated basal cell (PA) are marked by Roman numerals (I, II). Their fate is reported by morphology, distance from the basement membrane, and mitotic potential (a, b indicate 2nd generation progeny). Scale bar, 25 μm. d, Symmetric renewal is significantly suppressed in ubiquitously activated Pik3ca 2X epidermis. Statistics based on n=3 animals of each condition. 96 wild-type and 102 Pik3ca 2X divisions were scored. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. e and f, Schematic (e) and representative image (f) of live imaging of single CreER-transduced mT/mG cells activated by tamoxifen at P19 produced three modes of fate choice 48hrs post activation. Scale bar, 25 μm. g, Progenitor cell renewal is significantly suppressed in activated Pik3ca 2X epidermis. Statistics based on n=3 animals of each condition. 296 wild-type and 317 Pik3ca 2X doublets were scored. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Statistical source data for d,g are shown in Supplementary Table 4.
To investigate if oncogene-driven differentiation is a cell autonomous process, we transduced Cre-reporter mice with LV-CreER and treated them with tamoxifen at P19 (Fig. 3e). We confirmed that low-dose of the drug resulted in sporadic labeling of epidermal progenitors, and efficient activation of oncogenic Pik3ca expression (Supplementary Fig. 5c-h). At 48hrs, we observed that most labeled progenitors had divided and scored their daughter cell fates (Fig. 3f). As with ubiquitous oncogene activation, symmetric renewal was significantly reduced and both symmetric differentiation and asymmetric divisions significantly increased in Pik3ca 2X cells compared to wild-type (Fig. 3g).
Using three independent differentiation assays we established that progenitor renewal rate in Pik3ca 2X epidermis is significantly lower than in the wild-type (0.52 vs. 0.43; Supplementary Fig. 5i), and we expected this difference to have a profound effect on tissue growth12. Indeed, clonal expansion in epidermis under constant proliferation and tissue turnover rate (long-term EdU chase and41) is predicted to vary between homeostasis and rapid expansion or loss when renewal rates change by as little as 5–10% (Supplementary Fig. 6a,b). To test if oncogenic Pik3ca-driven differentiation is sufficient to promote loss of clonal expansion, we generated a model of clonal growth using proliferation and renewal rates observed in wild-type and Pik3ca 2X epidermis (Supplementary Fig. 6c). Our model predicted that single cell activation of oncogenic Pik3ca in the adult epidermis would result in clone loss, which was consistent with our long-term lineage tracing analysis (Fig. 4a,b). To test if oncogene-induced differentiation can affect tumor initiation, we transduced wild-type and Pik3ca 2X epidermis with lentivirus expressing HPV-E7 oncogene. We found that progenitor renewal and clonal expansion were significantly increased in HPV-E7 transduced wild-type epidermis, and that tumor growth was initiated as early as 1 month (Fig 4c-f). Importantly, oncogenic Pik3ca significantly suppressed progenitor renewal, clonal expansion and delayed tumor initiation driven by HPV-E7 (Fig 4c-f), which suggests that constitutively activated PI3K signaling can act as a tumor suppressor during epidermal tumor initiation.
Fig. 4. Oncogenic PI3K activation results in long-term clonal loss and suppresses tumor initiation.
a, Cre-inducible expression of oncogenic Pik3caH1047R is achieved in cells transduced with lentivirus for Tamoxifen (TAM)-dependent activation of Cre recombinase. Skin epidermis treated with TAM at postnatal (P) day 19 was imaged at P20, 21, 35, 42 and P105 to quantify clone size in wild-type and Pik3ca 2X epidermis. Pik3ca 2X clones are significantly smaller then wild-type at P35 and P42, and are lost from the tissue by P105. Statistics are based on n=3 animals of each condition, clone numbers for each day/condition: WT (P20:101, P21:84, P28:134, P35:129, P42:135, P105:93); Pik3ca 2X (P20:142, P21:127, P28:170, P35:88, P42:89). Two-tailed t test, P-value as indicated. Box 10–90 percentile, error bar: SD, center value: mean. b, Representative image of wild-type (WT) tissue and Pik3ca 2X clones in P105 epidermis. H2B-GFP (green nuclear staining) was used as transduction control. Scale bar, 100 μm. 90 WT regions and 93 Pik3ca 2X clones were imaged. c, Oncogenic Pik3ca suppresses rate of renewal in HPV E7 oncogene transduced epidermis and tumor. Statistics based on n=3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. d and e, Representative images (d) and statistics (e) show that oncogenic Pik3ca suppresses clonal expansion driven by HPV E7 oncogene. Statistics were derived from 36 WT+LV-HPV-E7 and 28 Pik3ca 2X+LV-HPV-E7 clones pooled from 3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Scale bar, 100 μm. f, Kaplan–Meier survival curve indicates oncogenic Pik3ca significantly delays HPV E7 driven tumor initiation. Statistics are based on n=8 animals of each condition. Log-rank test, P-value=0.0002. Statistical source data for a,c,e are shown in Supplementary Table 4.
AKT substrate SH3RF1 mediates PI3K-driven differentiation.
To functionally dissect the molecular mechanism of oncogenic Pik3ca-driven differentiation, we focused on AKT as the dominant effector kinase of PI3K signaling, which is activated in Pik3ca 2X epidermis (Supplementary Fig.2b). We selected shRNAs that efficiently deplete Akt1, Akt2, or Akt3 transcripts, and showed that silencing of Akt1 and Akt2, but not Akt3, significantly promoted progenitor renewal, in both wild-type and Pik3ca 2X tissues (Supplementary Fig. 7a-c). Akt1 and Akt2 were similarly required for oncogenic Pik3ca-driven proliferation (Supplementary Fig. 7d,e). To test if progenitor differentiation is regulated by a specific AKT substrate and independently of proliferation, we pooled 1062 shRNAs targeting 242 validated AKT substrates42, and performed two genetic screens in vivo (Fig. 5a). For the differentiation screen, we collected basal (α6 Itghigh) and suprabasal (α6 Itglow) epidermal cells (Fig. 5a). For the proliferation screen, we isolated dividing (EdU+) and non-dividing (EdU-) basal cells following an 8hr EdU pulse (Fig. 5a). We expected that: (i) shRNAs that inhibit differentiation drivers will be enriched in basal cells (pro-renewal; Fig. 5a,b); (ii) shRNAs against drivers of progenitor renewal will be enriched in suprabasal cells (pro-differentiation; Fig. 5a,b); and, (iii) shRNAs that are dispensable for proliferation will be equally present in EdU+ and EdU- progenitors (proliferation neutral; Fig. 5a,c).
Fig. 5. Genetic screens identify SH3RF1 as a specific mediator of oncogenic PI3K/AKT-induced differentiation but not proliferation.
a, Schematic of in vivo genetic screens for regulators of progenitor differentiation and proliferation among 242 known AKT substrates. Assays are based on enrichment (orange) or depletion (blue) of shRNAs in a6-Integrinhigh basal progenitors relative to differentiated suprabasal a6-Integrinlow cells (differentiation screen) or non-dividing EdU- relative to dividing EdU+ epidermal progenitors (proliferation screen). b-d, In vivo differentiation (b) and proliferation (c) screens identify Sh3rf1 as the top-ranking promoter of progenitor renewal that does not affect cell division (d). Fold change is an average value from n=3 biological replicates of each genetic screen. Significance quantified by Deseq2, with cutoff P-value=0.01. e and f, Immunoprecipitation of endogenous (e) and V5-tagged SH3RF1 (f) from epidermal lysates followed by Western blotting using phosphor-AKT-substrate specific antibody shows that SH3RF1 is phosphorylated by AKT in epidermis, and that its phosphorylation status is enhanced in Pik3ca 2X tissue. Experiment was repeated 3 times independently with similar results. Unprocessed blots see Supplementary Fig. 8. Depletion of Sh3rf1 in wild-type epidermis promotes progenitor differentiation without an effect on cell division rate, n=3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. h, Overexpression of Sh3rf1 inhibits oncogenic Pik3ca-driven differentiation (left) without an effect on the rate of cell division (right, n=3 animals of each condition). Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. i and j, Representative image (i) and quantification (j) shows that overexpression of SH3RF1 promotes clone growth in wild-type and rescues clone loss following oncogenic PI3K activation. Statistics were derived from n=90 clones pooled from 3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Scale bar, 100 μm. Statistical source data for g,h,j are shown in Supplementary Table 4.
We quantified shRNA representation in each group of cells36, and identified Sh3rf1 as the top candidate whose silencing promoted differentiation without an effect on proliferation (Fig. 5b-d, Supplementary Tables 2 and 3). To test if SH3RF1 is an AKT substrate in the epidermis, we pulled down endogenous protein, and probed it using phospho-AKT-substrate specific antibody. We detected AKT phosphorylation of SH3RF1 in wild-type epidermis that was further increased in Pik3ca 2X tissue (Fig. 5e). We observed the same when full-length SH3RF1 (SH3RF1WT) was expressed from a lentivirus (Fig. 5f), confirming that AKT phosphorylates SH3RF1 in vivo. To functionally test SH3RF1 as a specific differentiation regulator we performed a series of gain- and loss-of function experiments in vivo. To model AKT phosphorylation-mediated suppression of SH3RF1, we transduced wild-type epidermis with shRNA constructs that most efficiently depleted its transcript (Supplementary Fig. 7f). Sh3rf1 depletion promoted progenitor differentiation without significant effect on cell division, thus validating our genetic screens (Fig. 5g). To counter the effect of AKT phosphorylation, we overexpressed SH3RF1WT in Pik3ca 2X epidermis (Fig. 5h). To control for possible reduction in AKT phosphorylation that may be brought by substrate overexpression, we also expressed a known AKT substrate H3F3B. We observed that SH3RF1 expression, but not H3F3B, suppressed oncogenic Pik3ca-driven differentiation while proliferation was unchanged (Fig. 5h). Lastly, to test if SH3RF1 can regulate oncogene-driven clonal expansion, we overexpressed SH3RF1WT in wild-type and Pik3ca 2X epidermis and observed that the resultant clones were significantly larger and maintained long-term compared to Pik3ca 2X epidermis alone (Fig. 5i,j). Together, our data suggest that SH3RF1 is an AKT substrate in the epidermis, where it acts as a specific mediator of oncogenic Pik3ca-driven differentiation and clonal growth.
Activated PI3K/AKT inhibits SH3RF1-mediated support of JNK signaling to promote progenitor differentiation.
Sh3rf1 is an SH3 domain-containing scaffold molecule necessary for JNK signaling43. Sh3rf1 can also function as an E3 ubiquitin ligase to regulate additional pathways44,45. Importantly, the scaffold function of Sh3rf1 is independent of its E3 ligase activity, and Sh3rf1 with deleted RING finger domain (SH3RF1ΔRing) can still support JNK signaling46,47. In addition, Akt-mediated phosphorylation of Sh3rf1 only disrupts it’s scaffold function, and ability to activate JNKs43,46–49. Therefore, we hypothesized that Sh3rf1 function in oncogene-driven differentiation depends on its scaffold function in regulation of JNK signaling. We generated an allelic series of Sh3rf1 constructs (Fig. 6a,b), and expressed them in wild-type and Pik3ca 2X epidermis. We found that expression of SH3RF1ΔRing was equal to SH3RF1WT in promoting progenitor renewal and clonal expansion independently of proliferation (Fig. 6c-e, Supplementary Fig. 7g), confirming that E3 ubiquitin ligase function of SH3RF1 is dispensable in oncogene-induced differentiation.
Fig. 6. AKT-mediated phosphorylation of SH3RF1 is required for oncogenic Pik3ca-induced differentiation and suppression of clonal growth.
a, Schematic of SH3RF1 mutation constructs: deletion of Ring domain, SH3RF1ΔRing; AKT phosphorylation site mutant, SH3RF1SA; AKT phosphorylation mimetic mutant, SH3RF1SD. b, Immunoprecipitation of V5-tagged SH3RF1 mutants from transduced epidermis lysate followed by Western blotting using phosphor-AKT-substrate specific antibody confirmed that S304 is phosphorylated by AKT in epidermis. Experiment was repeated in n=3 biological replicates with similar results. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Unprocessed blots see Supplementary Fig. 8. c, EdU-BrdU pulse-chase differentiation assay shows that SH3RF1ΔRing was equal to SH3RF1WT in promoting renewal, and that SH3RF1SA further enhances renewal while SH3RF1SD does not, in both wild-type and Pik3ca 2X epidermis. Statistics are based on n=3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. d and e, Representative image (d) and quantification (e) shows that SH3RF1ΔRing was no different from SH3RF1WT in promoting clonal growth, and that SH3RF1SA but not SH3RF1SD further enhance clonal growth, in both wild-type and Pik3ca 2X epidermis. Statistics are based on 90 clones from n=3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Scale bar, 100 μm. Statistical source data for b,c,e are shown in Supplementary Table 4.
We next expressed AKT phosphorylation site (S304) mutated SH3RF1SA as well as phosphomimetic SH3RF1SD in wild-type and Pik3ca 2X epidermis (Fig. 6a,b). We observed that SH3RF1SD had no effect on progenitor renewal or clonal growth, while SH3RF1SA significantly promoted both processes (Fig. 6c-e, Supplementary Fig. 7g). These observations suggest that clonal expansion and progenitor renewal-promoting ability of SH3RF1 is dependent on, and suppressed by, AKT-mediated phosphorylation.
SH3RF1 promotes JNK signaling as a scaffold for assembly of MLK/MKK complex46,49. This process has been demonstrated to be negatively regulated by AKT mediated phosphorylation on S304 of SH3RF146,49. Meanwhile, JNK has been implicated as a positive regulator of progenitor renewal in skin50,51. Together, this suggested a molecular pathway, where AKT phosphorylation and suppression of SH3RF1 inhibits JNK signaling-mediated maintenance of progenitor cell fate (Fig. 7a). Consistent with our model, GSEA of the oncogenic Pik3ca transcriptome demonstrated a significant suppression of JNK signature genes compared to wild-type (Fig. 7b). Moreover, immunofluorescence staining of epidermal tissues showed that phospho-JNK was significantly reduced in basal progenitors expressing oncogenic Pik3ca, and that the staining is largely recovered upon SH3RF1WT expression (Fig. 7c, Supplementary Fig. 7h). To directly test if PI3K/AKT-mediated phosphorylation of SH3RF1 can modify epidermal JNK signaling, we performed a series of experiments. Western blot analyses showed that JNK phosphorylation is suppressed in Pik3ca 2X epidermis relative to wild-type, and that this suppression can be rescued by overexpression of SH3RF1WT (Fig. 7d). As SH3RF1ΔRing also rescued JNK phosphorylation, and expression of SH3RF1SA but not SH3RF1SD rescued and further enhanced phospho-JNK level in Pik3ca 2X epidermis, it suggested that Sh3rf1 promotes JNK signaling independently of its E3 ubiquitin ligase function, and that this positive regulation can be suppressed by Akt phosphorylation (Fig. 7e).
Fig. 7. SH3RF1 is a scaffold for JNK signaling in oncogenic Pik3ca-driven differentiation.
a, Model of how AKT signaling can promote epidermal progenitor cell differentiation. b, Transcriptome of Pik3ca 2X progenitors shows significant suppression of JNK signature genes. ES, NES, FDR generated by GSEA2 program. Statistics are based on RNA-seq performed in n=3 animals of each condition. c, p-JNK staining in indicated samples, dashed line indicates basement membrane. Scale bar, 25 μm. d, Western blotting and quantification of JNK phosphorylation in WT and Pik3ca 2X epidermis. Significant reduction of phospho-JNK signal in Pik3ca 2X epidermis is rescued by SH3RF1 expression. Experiment was repeated in n=3 biological replicates with similar results. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Unprocessed blots see Supplementary Fig. 8. e, SH3RF1ΔRing and SH3RF1WT but not SH3RF1SD rescue phospho-JNK suppression in Pik3ca 2X epidermis, while SH3RF1SA can further enhance it. Experiment was repeated in n=3 biological replicates with similar results. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Unprocessed blots see Supplementary Fig. 8. f and g, MLK1 rescues oncogenic Pik3ca-induced suppression of phospho-JNK (f) and differentiation (g). Experiment was repeated in n=3 biological replicates with similar results (f). Statistics are based on n=3 animals of each condition (g). Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Unprocessed blots see Supplementary Fig. 8. h and i, Knockdown of Jnk1 blocks SH3RF1SA enhanced phospho-JNK (h) and progenitor renewal (i). Experiment was repeated in n=3 biological replicates with similar results (h). Statistics are based on n=3 animals of each condition (i). Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. Statistical source data for d-i are shown in Supplementary Table 4.
Lastly, we tested if suppression of JNK signaling is necessary and/or sufficient for oncogenic Pik3ca-induced differentiation. We overexpressed JNK activator43,48,52 MLK1 (MAP3K9) in Pik3ca 2X epidermis, and observed increased phospho-JNK levels (Fig. 7f) and suppression of oncogenic Pik3ca-induced differentiation with no effect on proliferation (Fig. 7g, Supplementary Fig. 7i). This supports a model where JNK signaling is downstream of oncogenic Pik3ca in regulation of epidermal progenitor renewal, and that its activation is sufficient to overcome oncogene-induced differentiation. To test if suppression of JNK is necessary for oncogene-induced differentiation, we expressed SH3RF1SA in Pik3ca 2X epidermis, while at the same time we depleted Jnk1 using shRNA knockdown (Fig. 7h). We observed that silencing of Jnk1 significantly abrogated the rescue effect of SH3RF1SA on oncogenic Pik3ca-induced differentiation without affecting cell division (Fig. 7i, Supplementary Fig. 7j), confirming that JNK signaling is not only required for, but is the dominant mediator of SH3RF1 function in regulating differentiation. Together, we demonstrate that AKT-mediated phosphorylation of SH3RF1 and subsequent suppression of JNK signaling is a critical pathway in oncogenic Pik3ca-induced differentiation, and provide a mechanism for oncogenic tolerance without triggering cell cycle arrest or cell death in the skin epithelium.
Discussion
How tissues respond to oncogenic mutations to prevent clonal expansion has been a central question in cancer research for decades8,53,54. Senescence and apoptosis can block oncogene-driven clonal expansion in culture7,55, and senescence has been demonstrated to have a robust tumor suppressive potential under certain physiological scenarios4,5,56. However, both of these cellular processes feature an abrupt block to proliferation and tissue disruption, which is inconsistent with evidence that skin epithelium, despite a large quantity of oncogenic lesions3, can maintain both its proliferative and functional integrity. Thus, it is likely that skin, which depends on high rates of tissue turnover to maintain an efficient barrier to the external environment, employs a tumor suppressive mechanism that can maintain proliferation and also manage long-term expansion of oncogene-activated cells. Oncogenic Pik3ca-induced differentiation that we describe here fits these requirements as it allows the epidermis to manage oncogenic stress by driving excess oncogene-activated cells out of the mitotic progenitor pool.
Even though linage choice in epidermis is relatively simple, and differentiation is accompanied by observable and well-documented changes in cell morphology and marker expression, methods to directly access cell fate choice in vivo have been slow to emerge. Indeed, investigation of how oncogenes affect cell fate choices, and our discovery of oncogenic Pik3ca-induced differentiation was made possible only recently, through development of direct and quantitative assays, including intravital imaging41. In the current study, we add to this experimental toolbox by describing a simple EdU-BrdU pulse-chase assay as a sensitive method to quantify progenitor cell renewal rate in skin epithelium.
Animal studies have already shown that activation of the PI3K/AKT pathway in mice through oncogenic mutations in Pik3ca31,32 or loss of Pten57,58 does not result in epidermal tumorigenesis59, although tumors are initiated in multiple other epithelial organs. These observations suggest that the ability of oncogenic Pik3ca to promote tumorigenesis may be stringently restricted by a tumor suppressive mechanism in distinct epithelia. Curiously, differentiation that is accompanied by loss of proliferative potential is not seen across all epithelia, but seems specific for highly proliferative ones, such as skin epidermis60,61, intestinal epithelium62, and oral mucosa11. Future studies should explore if these tissues employ differentiation to block clonal expansion and counter oncogenic stress.
Our identification of differentiation as the dominant growth suppressive mechanism in skin epithelium further suggests that its loss in the context of oncogenic lesions would provide oncogene-activated clones with the ability to expand, while retaining higher proliferation rates at the same time. Supporting this notion, mutations identified as prominent clonal expansion promoters in both human tissue3 and our genetic screen, are always gained earlier in SCC progression than oncogenic lesions in PIK3CA63.
Methods
Animals.
We used equal numbers of male and female animals throughout the study. Pik3caH1047R/H1047R (donated by Wayne A. Phillips31–33), K14-H2B-PAmCherry (donated by Valentina Greco41), and R26mT/mG and R26yfp/yfp Cre-reporter mice (Jackson Laboratories) were on the C57BL/6 or C57BL/6-Tyrc−2J/J background. Mice were housed and cared for in an AAALAC-accredited facility at Fred Hutchinson Cancer Research Center, and all animal experiments were conducted in accordance with Fred Hutchinson Cancer Research Center ethical regulations and IACUC-approved protocols, project license number 50814.
Lentivirus production and transduction.
Large-scale production and concentration of lentivirus were performed as previously described26. Detailed description of the lentiviral transduction of 293T (Invitrogen) and primary keratinocytes in culture, and of in utero-guided lentiviral transduction in vivo can be found elsewhere26,27,64.
Lentiviral constructs.
ORF overexpression was achieved using pLX EF1 Barcode vector (Supplementary Fig. 1b). The pLX EF1 Barcode vector was modified from pLX302 (Addgene), in which we replaced: i) PGK-puroR cassette between PpuM1/Xba1 sites with a randomized 10bp barcode, ii) CMV promoter between Xho1/Nde1 sites with an EF1α promoter from pEF-BOS (Addgene). RNAi-mediated gene depletion was achieved using pLKO1 shRNA vectors from the mouse TRC1.0 shRNA library (Sigma-Aldrich). To construct the lentiviral pools and ensure equal lentivirus representation, plasmids were mixed together in equal molar ratios. Expression of Cre recombinase in tandem with ORF expression was achieved using pLX EF1 Cre vectors. This vector was modified from pLX302 (Addgene), by inserting Cre downstream of PGK promoter using Kpn1/Xba1 sites. Cre-shRNA expression was achieved using pLKO-Cre vectors described before26,27.
In vivo genetic screens.
To quantify construct abundance in the screen for clonal expansion, head skin of mice at P21 was collected and digested in 0.25% collagenase at 37 C° for 1 hour to release mesenchymal cells from the epithelium. Head skins from two animals were pooled and treated as a biological replicate, to achieve a ~200 fold coverage. To quantify construct abundance in the renewal and cell division screens, head skin of mice at E18.5 was digested in 2mg/ml dispase at 37 C° for 1 hour to separate epidermis from dermis. Epidermal tissue was further digested with 0.25% trypsin for 30 minutes into single cells. Head skins from 4 animals were pooled together to make one biological replicate, thus achieving a ~45 fold coverage. For the renewal screen, single epidermal progenitor cells were stained with CD326/EpCAM-APC (G8.8, 1:50; BD Biosciences) and CD49f/α6-Integrin-PerCP (GoH3, 1:50; Biolegend). For the cell division screen, single epidermal cells were subjected for Click-iT EdU detection (Invitrogen, C10338) followed by CD49f/α6-Integrin-PerCP staining. Cell populations of interest were isolated using BD FACSAria II machine (BD Biosciences). gDNA from all samples was extracted using QIAamp DNA tissue mini kit (Qiagen). Barcode pre-amplification, sequencing, and data processing using Deseq2 program65 were performed as previously described27,66.
Intravital imaging using 2-photon microscopy.
Intravital imaging was performed using LSM 780 multiphoton laser scanning confocal microscope (Zeiss) using previously described protocol41. Animal was immobilized using a modified custom-made device67 to minimize vibration from breathing during imaging of head skin. To photoactivate PA-H2B-mCherry, we exposed the cells for 2 minutes to continuous scan with 810nm laser set at 3%. Membrane-associated GFP and Tomato signals were captured using 940nm laser, PA-H2B-mCherry signal was captured using 1040nm laser.
Immunoprecipitation.
E18.5 epidermis was snap-frozen using LN2, mechanically pulverized, and lysed in lysis buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, and 1% Nonidet P-40). Lysates were incubated with anti-V5 affinity beads (Sigma-Aldrich, A7345) or dynabeads (Invitrogen, 10003D) pre-incubated with anti-SH3RF1 antibody (3H3, Abnova) overnight at 4°C. Beads with affinity-bound proteins were washed 6 times with immunoprecipitation wash buffer (200 mM NaCl, 10 mM HEPES, pH 7.4, and 0.1% Nonidet P-40) before direct loading onto a 4–12% gel and Western blot analysis.
Immunofluorescence and Western blot analyses.
The following primary antibodies were used: chicken anti-GFP (ab13970, 1:1000 for IF; Abcam); mouse anti-BrdU (MoBU-1, 1:100 for IF; Invitrogen); mouse anti-β-Actin (66009, 1:3000 for WB; Proteintech); mouse anti-V5 (V5–10, 1:3000 for WB; Sigma-Aldrich); rabbit anti-Keratin 10 (Poly19054, 1:1000 for IF; BioLegend); rabbit anti-Phospho-AKT (D9E, 1:1000 for WB; Cell Signaling); rabbit anti-total AKT (C67E7, 1:500 for WB; Cell Signaling); mouse anti-SH3RF1 antibody (3H3, 1:400 for WB; Abnova); rabbit anti-Phospho-AKT Substrate (110B7E, 1:1000 for WB; Cell Signaling); rabbit anti-Phospho-SAPK/JNK (81E11, 1:500 for WB, 1:100 for IF; Cell Signaling); rabbit anti-total SAPK/JNK (9252, 1:1000 for WB; Cell Signaling). Tissues were processed for immunostaining as previously described26,27, and mounted in ProLong Gold with or without DAPI (Life Technologies). For EdU-BrdU pulse-chase differentiation assay, tissue sections were first processed for EdU Click-iT following manufacturer’s instructions. Next, tissues were fully processed for K10 and GFP immunofluorescence detection. Lastly, tissue sections were incubated in 2N HCl at 37°C for 30 minutes to denature DNA, quenched with 0.1M sodium borate pH 8.5 twice, and processed for BrdU immunofluorescence detection. Confocal images were taken on a Zeiss LSM700 system, using Plan-Apochromat 40x/1.4 oil objective. Images were processed using Zeiss Zen and ImageJ software. Western blotting was performed using Novex system (Invitrogen) and chemiluminescent signal was captured using Odyssey Fc system (LI-COR).
EdU-BrdU cell division assay and quantification.
To measure the average cell cycle interval in epidermis, we developed a cell cycle assay based on specific labeling of cell that have completed cell division within a defined time frame. By using such method we minimized the potential artifact of conventional nucleotide incorporation based proliferation assay, i.e. tissue with longer S phase but not essentially faster division rate can appear to have more nucleotide incorporation (Supplementary Fig. 2d). To ensure the specificity of the assay, we tested BrdU specific antibody MoBU-1 and confirmed that animals injected with BrdU were readily labeled, while EdU (or sham) injected animals showed no staining. Similarly, we detected EdU using Click-iT chemistry only in animals injected with EdU.
To perform the assay, as illustrated in Supplementary Figure 2d, we first administered EdU to animals, followed by BrdU injection 2 hours later. We expected that cells that complete the S-phase during the first 2 hours would incorporate EdU only, while cells subsequently going through S-phase would incorporate both EdU and BrdU (Supplementary Fig. 2d). 6 hours later we harvest epidermis and detect EdU and BrdU signal by IF. At this time point we expect cells that complete the S-phase during the initial 2 hours and subsequently divided would give rise to 2 EdU+ only progenies while cells that stalled in G2 would only result in 1 EdU+ only cells (Supplementary Fig. 2d). However, we show that over our timeframe most cells divide (~97%), and that only a minority in both wild-type and Pik3ca 2X epidermis remains in G2 phase (Supplementary Fig. 3c,d).
To calculate cell division interval we define the number of EdU+ only cells as E. As one cell division gives rise to two daughter cells, the number of cell divisions that give rise to EdU+ only cells is E/2. As we have demonstrated, only basal cells have the potential to divide in the epidermis (Supplementary Fig. 3a,b); therefore, the fraction of basal cells that divide within two hours to give rise to EdU+ only cells is F=(E/2)/B; (B: total number of basal cells). We next extrapolate this measured to the whole basal cell population, by using the following ratio: D/1=2hrs/F, where D stands for the average cell division interval (in hours).
EdU-BrdU pulse-chase differentiation assay and quantification.
Since we can uniquely label a population of cells (EdU+ only) that had undergone S phase within the initial 2 hours and subsequently divided as described above, next, we set out to determine how long after cell division an average daughter cell takes before starting to express a differentiation marker K10. We administered EdU and then BrdU, and collected tissues every 2 hours. Tissues were processed to detect EdU and BrdU together with K10 (to mark committed progeny). Our analysis showed that, K10 expression was detected as early as 2 hours, and plateaued by 6 hours of BrdU labeling. Importantly, similar dynamics were observed in both WT and Pik3ca 2X tissues, and irrespective of which nucleoside analog was administered first. Thus by analyzing epidermis 6 hours after the start of BrdU labeling, we can calculate the rate of progenitor cell renewal of the EdU+ only cells using the following equation: rate of renewal= number of EdU+ only K10- cells / (total number of EdU+ only cells).
RNA-seq and GSEA analyses.
We isolated inter-follicular epidermis from dermis and hair follicle using dispase digestion. Single cells were obtained following 15 minute trypsin digestion, and stained for basal integrin expression64. We FACS isolated α6-Integrinhigh epidermal progenitors TrizolLS (Invitrogen), extracted their RNA using phenol/chloroform protocol, and purified it using QIAamp RNA mini kit (Qiagen), per manufacturer instructions. RNA quality was assessed using Agilent 2100 Bioanalyzer, with all samples passing the quality threshold of RNA integrity numbers (RIN)>8. Library was prepared using Illumina TrueSeq mRNA sample preparation kit at the Fred Hutchinson Cancer Research Center Genomic Core facility, and cDNA was sequenced on Illumina HiSeq 2000. Reads were mapped to mm9 build of the mouse genome using TopHat, and transcript assembly and differential expression were determined using Cufflinks68. Gene Set Enrichment Analysis69 was performed using GSEA2. Reference gene expression signatures were obtained from previously published expression profiles: HFSC34, mouse basal/suprabasal cells35, human basal/suprabasal cells36, JNK activation signature70.
Statistics and Reproducibility.
All quantitative data were expressed as mean ± s.d. Differences between groups were assayed by two-tailed student t-test or one-way ANOVA using Prism 5 (GraphPad software). Significant differences were considered when P<0.05. All quantitative data were collected from experiments performed in at least three samples or biological replicates. The sample size was not predetermined and the experiments were not randomized.
Supplementary Material
Acknowledgements
We thank W. Phillips for sharing the inducible Pik3caH1047R mouse; V. Greco and C. Pineda for sharing photoactivatable mouse and the intravital imaging technique; M. Schober and A. Hsieh for critical reading of the manuscript; Comparative Medicine (AAALAC accredited; R. Uthamanthil, director) for care of mice in accordance with National Institutes of Health (NIH) guidelines; Genomics (J. Delrow, director) for sequencing; Scientific Imaging (J. Vazquez, director) for advice; Flow Cytometry (A. Berger, director) for flow cytometry and FACS. This research was supported by grants from the NIH (R01-AR070780; S.B.), Cell & Molecular Biology Training Grant (M.S.), and Thomsen Family Fellowship (Z.Y.). We dedicate this work to Xiuting Chen, and are grateful for her strength and support.
Footnotes
All authors provided intellectual input, vetted and approved the final manuscript, and declare that they have no financial and non-financial competing interests.
Data availability. RNA–seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE99659. Previously published microarray data that were re-analysed here are available under accession code GSE4170434, GSE4885935, GSE2605936, GSE5053070.
Source data for Fig. 1e, 2b-d, 2f, 3d, 3g, 4a, 4c, 4e, 5g, 5h, 5j, 6c, 6e, 7g, 7i and Supplementary Fig. 2c, 3a, 3d, 4a-c, 4e, 5c-e, 5i, 7a-g, 7i, 7j have been provided as Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674, doi: 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Pardal R, Molofsky AV, He S & Morrison SJ Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harbor symposia on quantitative biology 70, 177–185, doi: 10.1101/sqb.2005.70.057 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Martincorena I et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886, doi: 10.1126/science.aaa6806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michaloglou C et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724, doi: 10.1038/nature03890 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Braig M et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665, doi: 10.1038/nature03841 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Chen Z et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730, doi: 10.1038/nature03918 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearnhead HO et al. Oncogene-dependent apoptosis is mediated by caspase-9. Proceedings of the National Academy of Sciences of the United States of America 95, 13664–13669 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe SW, Cepero E & Evan G Intrinsic tumour suppression. Nature 432, 307–315, doi: 10.1038/nature03098 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Bissonnette RP, Echeverri F, Mahboubi A & Green DR Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature 359, 552–554, doi: 10.1038/359552a0 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Fuchs E & Raghavan S Getting under the skin of epidermal morphogenesis. Nature reviews. Genetics 3, 199–209, doi: 10.1038/nrg758 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Jones KB & Klein OD Oral epithelial stem cells in tissue maintenance and disease: the first steps in a long journey. International journal of oral science 5, 121–129, doi: 10.1038/ijos.2013.46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasetti C & Levy D Role of symmetric and asymmetric division of stem cells in developing drug resistance. Proceedings of the National Academy of Sciences of the United States of America 107, 16766–16771, doi: 10.1073/pnas.1007726107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning BD & Cantley LC AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274, doi: 10.1016/j.cell.2007.06.009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zumsteg ZS et al. Taselisib (GDC-0032), a Potent beta-Sparing Small Molecule Inhibitor of PI3K, Radiosensitizes Head and Neck Squamous Carcinomas Containing Activating PIK3CA Alterations. Clinical cancer research : an official journal of the American Association for Cancer Research 22, 2009–2019, doi: 10.1158/1078–0432.CCR-15–2245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizrachi A et al. Tumour-specific PI3K inhibition via nanoparticle-targeted delivery in head and neck squamous cell carcinoma. Nature communications 8, 14292, doi: 10.1038/ncomms14292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins F, de Sousa SC, Dos Santos E, Woo SB & Gallottini M PI3K-AKT-mTOR pathway proteins are differently expressed in oral carcinogenesis. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology 45, 746–752, doi: 10.1111/jop.12440 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Watanabe S et al. Activation of PI3K-AKT pathway in oral epithelial dysplasia and early cancer of tongue. The Bulletin of Tokyo Dental College 50, 125–133 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Liu S et al. The PI3K-Akt pathway inhibits senescence and promotes self-renewal of human skin-derived precursors in vitro. Aging cell 10, 661–674, doi: 10.1111/j.1474–9726.2011.00704.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pankow S, Bamberger C, Klippel A & Werner S Regulation of epidermal homeostasis and repair by phosphoinositide 3-kinase. Journal of cell science 119, 4033–4046, doi: 10.1242/jcs.03175 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Calautti E, Li J, Saoncella S, Brissette JL & Goetinck PF Phosphoinositide 3-kinase signaling to Akt promotes keratinocyte differentiation versus death. The Journal of biological chemistry 280, 32856–32865, doi: 10.1074/jbc.M506119200 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Janes SM et al. PI3-kinase-dependent activation of apoptotic machinery occurs on commitment of epidermal keratinocytes to terminal differentiation. Cell research 19, 328–339, doi: 10.1038/cr.2008.281 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunschmann C et al. Insulin/IGF-1 controls epidermal morphogenesis via regulation of FoxO-mediated p63 inhibition. Developmental cell 26, 176–187, doi: 10.1016/j.devcel.2013.05.017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dainichi T et al. PDK1 Is a Regulator of Epidermal Differentiation that Activates and Organizes Asymmetric Cell Division. Cell reports 15, 1615–1623, doi: 10.1016/j.celrep.2016.04.051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segrelles C et al. Akt signaling leads to stem cell activation and promotes tumor development in epidermis. Stem cells 32, 1917–1928, doi: 10.1002/stem.1669 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Murayama K et al. Akt activation induces epidermal hyperplasia and proliferation of epidermal progenitors. Oncogene 26, 4882–4888, doi: 10.1038/sj.onc.1210274 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Beronja S, Livshits G, Williams S & Fuchs E Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nature medicine 16, 821–827, doi: 10.1038/nm.2167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beronja S et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501, 185–190, doi: 10.1038/nature12464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanhaesebroeck B, Stephens L & Hawkins P PI3K signalling: the path to discovery and understanding. Nature reviews. Molecular cell biology 13, 195–203, doi: 10.1038/nrm3290 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Li YY et al. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research 21, 1447–1456, doi: 10.1158/1078–0432.CCR-14–1773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzumdar MD, Tasic B, Miyamichi K, Li L & Luo L A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605, doi: 10.1002/dvg.20335 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Kinross KM et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. The Journal of clinical investigation 122, 553–557, doi: 10.1172/JCI59309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tikoo A et al. Physiological levels of Pik3ca(H1047R) mutation in the mouse mammary gland results in ductal hyperplasia and formation of ERalpha-positive tumors. PloS one 7, e36924, doi: 10.1371/journal.pone.0036924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hare LM et al. Physiological expression of the PI3K-activating mutation Pik3ca(H1047R) combines with Apc loss to promote development of invasive intestinal adenocarcinomas in mice. The Biochemical journal 458, 251–258, doi: 10.1042/BJ20131412 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Blanpain C, Lowry WE, Geoghegan A, Polak L & Fuchs E Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648, doi: 10.1016/j.cell.2004.08.012 (2004). [DOI] [PubMed] [Google Scholar]
- 35.De Craene B et al. Epidermal Snail expression drives skin cancer initiation and progression through enhanced cytoprotection, epidermal stem/progenitor cell expansion and enhanced metastatic potential. Cell death and differentiation 21, 310–320, doi: 10.1038/cdd.2013.148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radoja N, Gazel A, Banno T, Yano S & Blumenberg M Transcriptional profiling of epidermal differentiation. Physiological genomics 27, 65–78, doi: 10.1152/physiolgenomics.00031.2006 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Rendl M, Polak L & Fuchs E BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes & development 22, 543–557, doi: 10.1101/gad.1614408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adam RC et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370, doi: 10.1038/nature14289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams SE, Beronja S, Pasolli HA & Fuchs E Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353–358, doi: 10.1038/nature09793 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuchs E Epidermal differentiation and keratin gene expression. Journal of cell science. Supplement 17, 197–208 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Rompolas P et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474, doi: 10.1126/science.aaf7012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hornbeck PV et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic acids research 40, D261–270, doi: 10.1093/nar/gkr1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa C, Tarras S, Taylor J & Vojtek AB Akt2 negatively regulates assembly of the POSH-MLK-JNK signaling complex. The Journal of biological chemistry 278, 47922–47927, doi: 10.1074/jbc.M307357200 (2003). [DOI] [PubMed] [Google Scholar]
- 44.de Bock CE et al. Protein interaction screening identifies SH3RF1 as a new regulator of FAT1 protein levels. FEBS letters 591, 667–678, doi: 10.1002/1873–3468.12569 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Tuvia S et al. The ubiquitin E3 ligase POSH regulates calcium homeostasis through spatial control of Herp. The Journal of cell biology 177, 51–61, doi: 10.1083/jcb.200611036 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z, Kukekov NV & Greene LA POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. The EMBO journal 22, 252–261, doi: 10.1093/emboj/cdg021 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Z, Sproul A, Wang W, Kukekov N & Greene LA Siah1 interacts with the scaffold protein POSH to promote JNK activation and apoptosis. The Journal of biological chemistry 281, 303–312, doi: 10.1074/jbc.M509060200 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Lyons TR et al. Regulation of the Pro-apoptotic scaffolding protein POSH by Akt. The Journal of biological chemistry 282, 21987–21997, doi: 10.1074/jbc.M704321200 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Xu Z & Greene LA Activation of the apoptotic JNK pathway through the Rac1-binding scaffold protein POSH. Methods in enzymology 406, 479–489, doi: 10.1016/S0076–6879(06)06036–8 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Weston CR et al. The c-Jun NH2-terminal kinase is essential for epidermal growth factor expression during epidermal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America 101, 14114–14119, doi: 10.1073/pnas.0406061101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang JY, Green CL, Tao S & Khavari PA NF-kappaB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes & development 18, 17–22, doi: 10.1101/gad.1160904 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Z, Maroney AC, Dobrzanski P, Kukekov NV & Greene LA The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Molecular and cellular biology 21, 4713–4724, doi: 10.1128/MCB.21.14.4713–4724.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greaves M & Maley CC Clonal evolution in cancer. Nature 481, 306–313, doi: 10.1038/nature10762 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marshall CJ Tumor suppressor genes. Cell 64, 313–326 (1991). [DOI] [PubMed] [Google Scholar]
- 55.Serrano M, Lin AW, McCurrach ME, Beach D & Lowe SW Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Munoz-Espin D & Serrano M Cellular senescence: from physiology to pathology. Nature reviews. Molecular cell biology 15, 482–496, doi: 10.1038/nrm3823 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Podsypanina K et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proceedings of the National Academy of Sciences of the United States of America 96, 1563–1568 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li G et al. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development 129, 4159–4170 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Du L et al. Overexpression of PIK3CA in murine head and neck epithelium drives tumor invasion and metastasis through PDK1 and enhanced TGFbeta signaling. Oncogene 35, 4641–4652, doi: 10.1038/onc.2016.1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuchs E Scratching the surface of skin development. Nature 445, 834–842, doi: 10.1038/nature05659 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watt FM, Lo Celso C & Silva-Vargas V Epidermal stem cells: an update. Current opinion in genetics & development 16, 518–524, doi: 10.1016/j.gde.2006.08.006 (2006). [DOI] [PubMed] [Google Scholar]
- 62.van der Flier LG & Clevers H Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annual review of physiology 71, 241–260, doi: 10.1146/annurev.physiol.010908.163145 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Hao JJ et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nature genetics 48, 1500–1507, doi: 10.1038/ng.3683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beronja S & Fuchs E RNAi-mediated gene function analysis in skin. Methods in molecular biology 961, 351–361, doi: 10.1007/978–1-62703–227-8_23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550, doi: 10.1186/s13059–014-0550–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schramek D et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343, 309–313, doi: 10.1126/science.1248627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu X & Zuo Y Two-photon in vivo imaging of dendritic spines in the mouse cortex using a thinned-skull preparation. Journal of visualized experiments : JoVE, doi: 10.3791/51520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology 28, 511–515, doi: 10.1038/nbt.1621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gozdecka M et al. JNK suppresses tumor formation via a gene-expression program mediated by ATF2. Cell reports 9, 1361–1374, doi: 10.1016/j.celrep.2014.10.043 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.