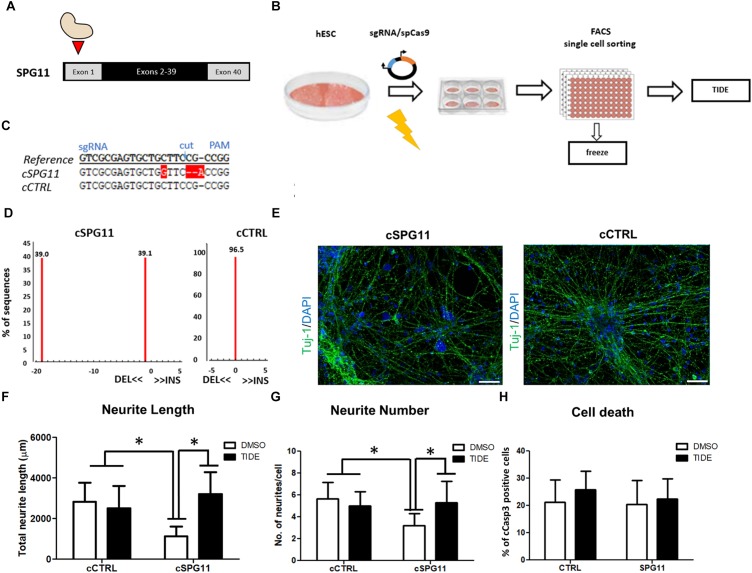
FIGURE 4.
CRISPR/Cas9 genome edited hESC-derived-neurons recapitulate the neuritic phenotypes of patient iPSC-derived-neurons. (A) Strategy for SPG11 knock-out by targeting exon 1 of SPG11. (B) Experimental scheme for CRISPR/Cas9 mediated genome editing of hESCs. hESCs were nucleofected with 5 μg of pX330 plasmid expressing SpCas9 and the sgRNA. After 48-h, the cells were single cell-sorted into 96-well plate. Subsequently, emerging colonies were split in 1:2 ratio. Half of the split was used for genomic DNA generation. The rest was transferred into a new 96-well plate for cryopreservation in –80°C. For identification of positive clones, PCR products were sequenced, further expanded and genotyped by Tracking Indels by Decomposition (TIDE; https://tide.nki.nl/) web-tool. (C) Sequence comparison between reference non-nucleofected HUES6 cell line, a positive, genome edited clone (cSPG11) and a control that underwent the genome editing process, but had no mutations (cCTRL). (D) TIDE analysis of cSPG11 and cCTRL. (E) Representative images of the genome edited lines stained for Tuj-1 (green). (F) The total neuritic length is significantly decreased in cSPG11 neurons and it is recovered following tideglusib treatment. (G) The average number of SPG11 neurites is significantly decreased and is recovered following tideglusib treatment. (H) Quantification of cCasp3+ cells reveals no significant difference between cSPG11 and cCTRL. Data shown as means ± SD. Scale bar = 50 μm. ∗P < 0.05. FACS = Fluorescence-activated cell sorting.