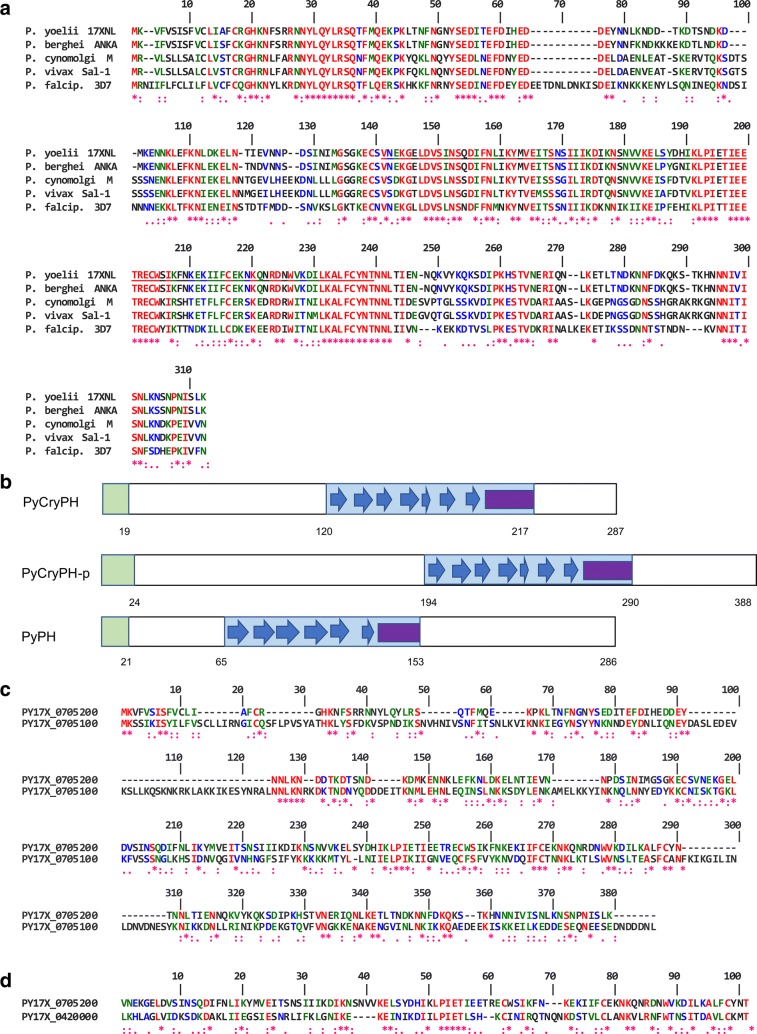
Fig. 1.
Sequence alignment and predicted secondary structure of CryPH compared with its orthologues and PH domain family proteins. a Alignment of amino acid sequences of CryPH orthologues in Plasmodium. Amino acid sequences of Plasmodium CryPH (PY17X_0705200, PBANKA_0704900, PcyM_0507700, PVX_089075, PF3D7_0825700) are aligned using CLUSTALW program (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html). b Schematic representation of the secondary structure of PyCryPH, PyCryPH-p (PY17X_0705100), and PyPH (PY17X_0420000). Green and blue squares indicate the N-terminal signal peptide and PH domain. Blue arrows and purple square indicate ß-sheets and α-helix. Numbers indicate amino acid positions from the first methionine. c Amino acid sequence comparison between PyCryPH and PyCryPH-p. Twenty percent of residues including all four cysteine residues are conserved between PyCryPH and PyCrePH-p. d Amino acid sequence alignment between PH domains of PyCryPH and PyPH. “*”, “:”, and “.” indicate amino acid residues with full identity, strong similarity, and weak similarity, respectively