Fig. 6.
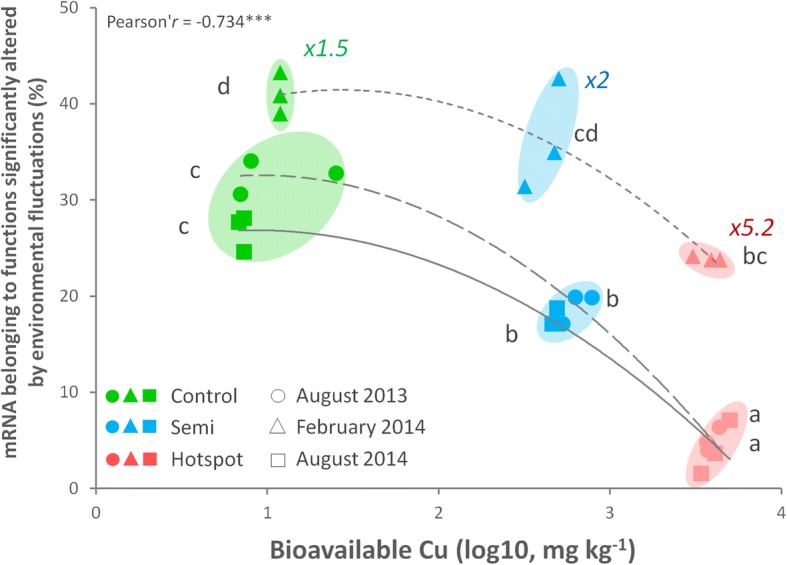
Cu-mediated decoupling of the temporal microbial mRNA response. The x-axis gives the bioavailable Cu quantified in each soil samples. The y-axis represents the sum of mRNA sequences in each corresponding metatranscriptomic sample belonging to SEED functions that were significantly altered by environmental fluctuation via pairwise comparison between time points (in percentage of total annotated sequences, excluding phages). The decrease of temporally affected mRNA functions along increasing bioavailable Cu (log10, mg kg−1) was inferred using the Pearson correlation coefficient (r = − 0.73***, p = 2.0E−5). Statistical differences between time points in each plot were inferred with ANOVA (Tukey’s HSD post hoc test, p < 0.05). Letters are attributed in ascending order, “a” being the smallest average. Different letters indicate statistically significant differences (p < 0.05). The colored numbers indicates the observed increase folds in mRNA sequences counts in February 2014 against both August time points
