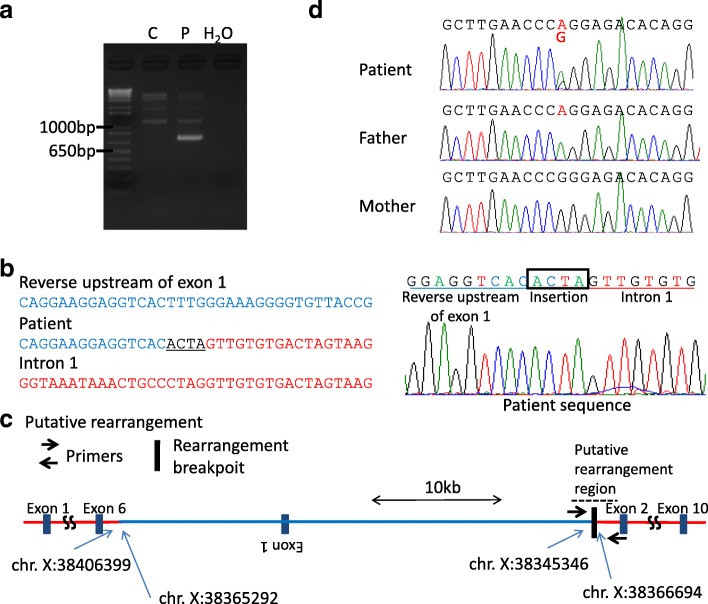
Fig. 3.
The complex rearrangement in the OTC gene of the study patient likely occurred via a replication-based mechanism. a Identification of the other junction by PCR. The PCR primer pair successfully amplified the junction product only from the OTCD study patient DNA. P, patient; C, control; H, H2O. b Sanger sequencing results for the PCR products including the junction. The normal sequence of the OTC gene upstream region and intron 1 are aligned in blue and red typeface, respectively. Underlined bases denote a non-templated microinsertion at the junction. c Predicted structure of the complex rearrangement leading to the OTC gene duplication. The positions of the PCR primers are indicated by black arrows. The first junction is indicated by a blue arrow. The nucleotide position of the breakpoints on the X-chromosome are also indicated. The position of the two breakpoints in intron 1 were found to be chr. X: 38365292 and chr. X: 38366694, which were 1402 bp apart. d Trio-genotyping of the common single nucleotide variant (rs752750694, NM 000531.5:c.-844C > T). The patient’s father carries A and her mother carries G/G. The patient was found to be an A/G heterozygote, but her A peak was two-fold higher than the G-peak. The areas under the curve (AUC) were 900 (A) and 328 (G) (Image J)