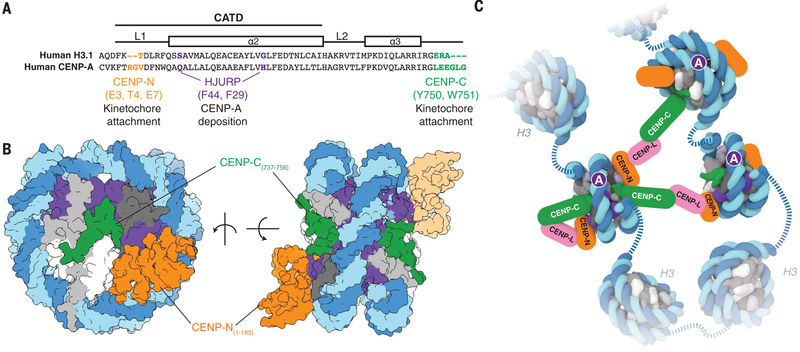
Fig. 4. Structural determinants of kinetochore assembly on the CENP-A nucleosome.
(A) Sequence alignment between humanH3.1 and CENP-A to highlight distinct CENP-A motifs involvedin deposition and recognition of CENP-A at centromeric chromatin (see also fig. S11, C and D). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (B) Two different views of the CENP-A nucleosome bound to hCENP-N and a modeled CENP-C motif peptide (5) to highlight potential dual binding of full-length CENP-C and CENP-N proteins on the CENP-A nucleosome. The second CENP-N (shown with lighter shading) is modeled on the basis of the cryo-EM density map obtained in the presence of excess hCENP-N1–286 (fig. S4), whereas the CENP-C motif peptides (human numbering shown for clarity) on each face of the nucleosome are positioned according to the crystal structure of the nucleosome in complex with the rat CENP-C motif (5). (C) Schematic view to highlight recognition and possible enrichment of CENP-A nucleosomes by the CCAN proteins CENP-C, CENP-N, and CENP-L. Other kinetochore proteins and the dimerization of CENP-C have been omitted for clarity.