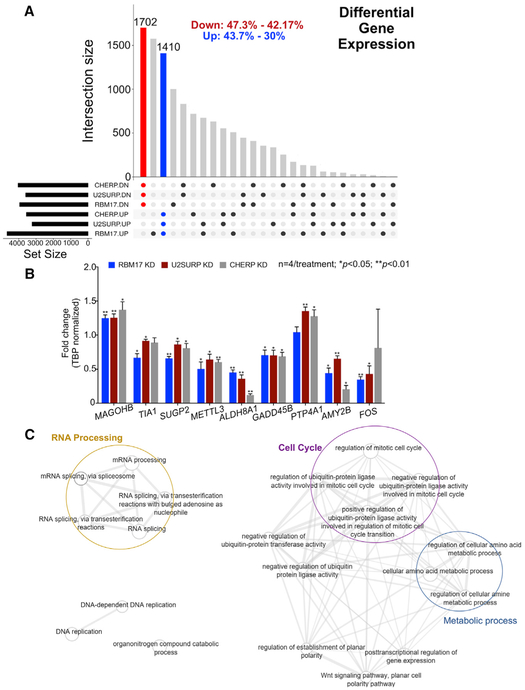
Figure 5. Knockdown of RBM17, U2SURP, or CHERP Alters Expression of RNA-Processing Factors.
(A) Plot showing the overlap pattern of differentially expressed genes across the knockdown models of RBM17, U2SURP, or CHERP (n = 3 replicates/siRNA, including siScramble). The red and blue bars represent all genes common to the three datasets (from the three interacting proteins)and consistently altered in the same direction: downregulated (red) and upregulated (blue). The black dots beneath the x axis indicate which proteins have been knocked down for each dataset and the direction of the resulting gene expression changes. The percentages at the top of the graph describe the size of the group of overlapping changes relatively to the biggest and the smallest of the three datasets.
(B) Validation of selected gene expression changes by qRT-PCR. The levels of selected transcripts were measured using qRT-PCR on samples from HEK293T cells treated with siRBM17, siU2SURP, or siCHERP and compared to cells treated with siScramble (n = 4 replicates/siRNA run in technical triplicates). Bars represent mean ± SEM; p value was calculated by two-tailed Student’s t test and significance was set at p < 0.05.
(C) Network plot of functionally enriched categories in overlapping differentially expressed genes. Each node is a functional category. The width of the connector between nodes is directly proportional to the number of genes shared between respective categories.