Figure 2.
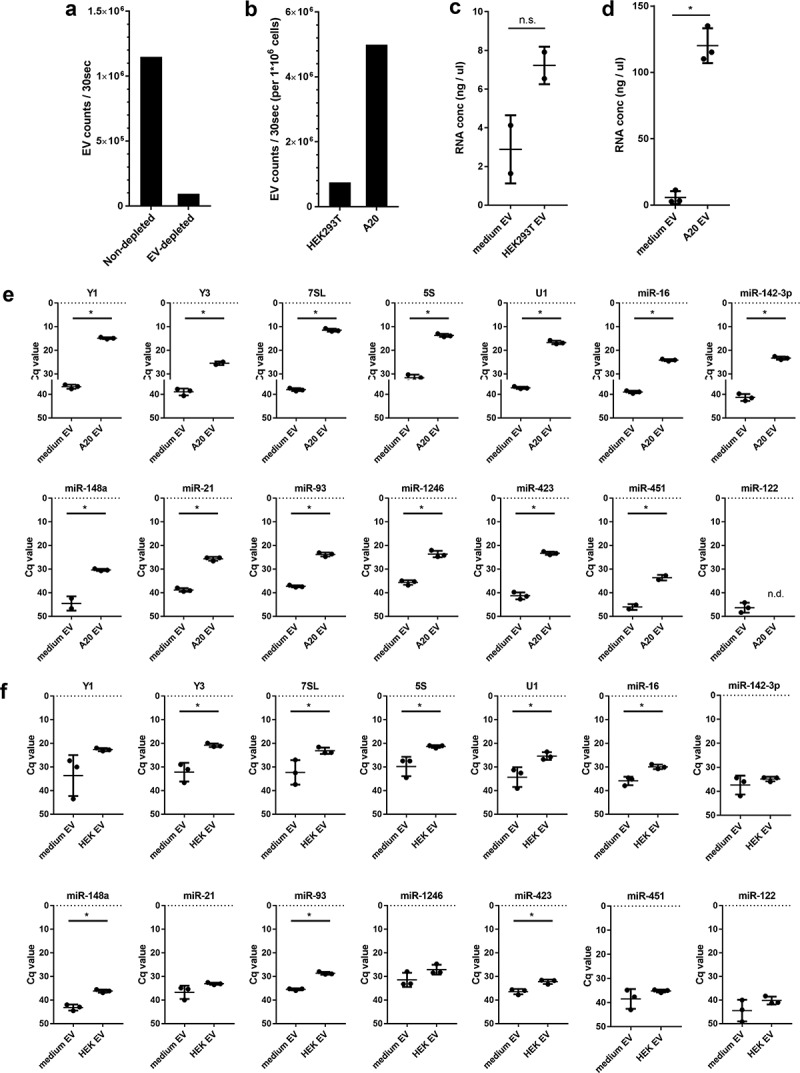
(a) EV-depleted medium was prepared using supernatant of ultracentrifuged solutions of 30% FCS. EV from equal volumes of EV-depleted and non-depleted medium were isolated by ultracentrifugation, fluorescently labelled with PKH67, and further purified by sucrose density gradient centrifugation. High-resolution flow cytometry was used to determine the number of PKH67-labeled EV. Indicated are the number of fluorescent EV detected in 30 s in pools of 1.12–1.18 g/mL sucrose fractions. (b) HEK293T and A20 cells were grown 20h in EV-depleted medium (prepared from 30% EV-depleted FCS). EV were isolated from equal volumes of HEK293T and A20 cell-conditioned supernatant, labelled with PKH67, and quantified using high-resolution flow cytometry. Indicated are the number of fluorescent EV in 30 s in pools of 1.12–1.18 g/mL sucrose fractions, normalised to the number of cultured cells to allow direct comparison of EV-release by both cell types. (c, d) HEK293T cells or A20 cells were cultured in EV-depleted medium (prepared from 30%-depleted FCS). EV were isolated from equal volumes of HEK293T (c) or A20 (d) conditioned medium and non-conditioned medium using differential centrifugation followed by sucrose density gradient centrifugation. RNA was isolated from EV pelleted from 1.12–1.18 g/mL sucrose fractions. Indicated are mean RNA concentrations ± SD of n = 2 (HEK293T) or n = 3 (A20) replicates. Statistical differences were determined by independent samples t-test, * p < 0.05. (E, F) EV-RNA isolated from A20 (e) and HEK293T (f) conditioned medium and equal volumes of non-conditioned EV-depleted medium (prepared as in d) were used for analysis of the indicated RNAs using RT-qPCR. Indicated are mean Cq values ± SD of n = 3 replicates. Statistical differences were determined by independent samples t-test, * p < 0.05.
