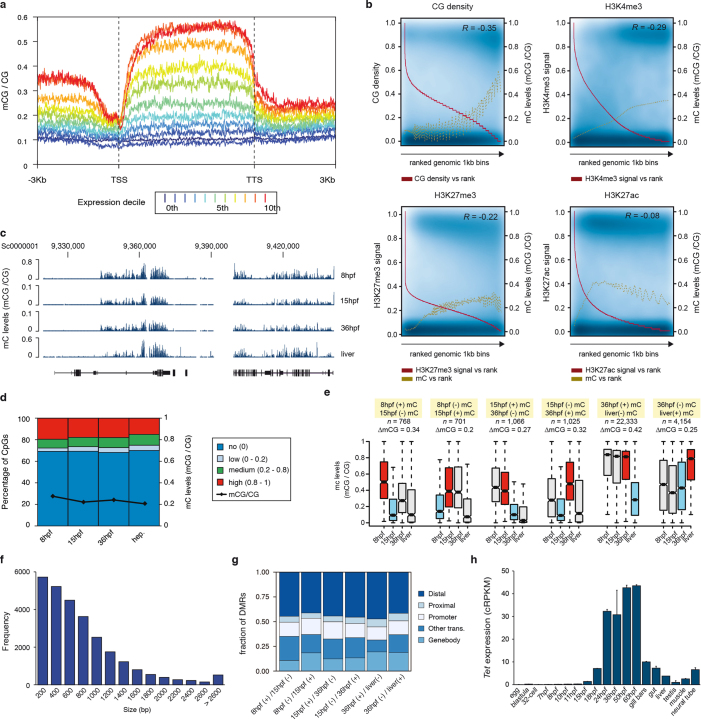
Extended Data Fig. 5. 5mC dynamics in amphioxus.
a, 5mC levels across gene bodies (n = 20,569) from different expression deciles (0th, not expressed; 10th, highest expression). TTS, transcription termination site. b, Scatter plots of levels of 5mC and CpG density, H3K4me3, H3K27me3 and H3K27ac in 1-kbp genomic bins sorted on the basis of feature rank. The red line tracks anti-correlation between feature density and rank number (a low rank number implies high feature density). The golden line represents a smoothing spline of 5mC signal versus feature rank number. Pearson correlation coefficients (R) are displayed in the top right corner of each panel. c, UCSC browser excerpt of 5mC patterns for selected regions. d, Percentage of methylated CpG dinucleotides in 8-hpf (n = 19,657,388), 15-hpf (n = 21,247,615), 36-hpf (n = 21,702,000) and hepatic (adult, n = 19,240,245) amphioxus samples. Black line indicates the fraction between methylated and non-methylated CpGs at each stage. e, Box plots of average 5mC levels in different types of differentially methylated regions (DMRs) at each stage. ΔmCG denotes the change in the fraction of methylated CpGs between the two stages used for identification of DMRs (red (hyper) and blue (hypo) boxes). The number of DMRs were as follows: 8 hpf(+)–15 hpf(−), 768; 8 hpf(−)–15 hpf(+), 701; 15 hpf(+)–36 hpf(−),1,066; 15 hpf(−)–36 hpf(+), 1,025; 36 hpf(+)–liver(−), 22,333; and 36 hpf(−)–liver(+), 4,154. The coordinates for all DMRs are provided in Supplementary Data 2, dataset 11. f, Distribution of DMR sizes (in bp). g, Genomic distribution of DMRs identified for each sample. ‘Other trans.’, DMRs that overlap with gene models that were not defined as being supported by orthology. h, Expression (cRPKMs) of the amphioxus Tet orthologue in embryos and adult tissues. Error bars represent standard error of the mean (the number of replicates for each RNA-seq dataset is provided in Fig. 1a).