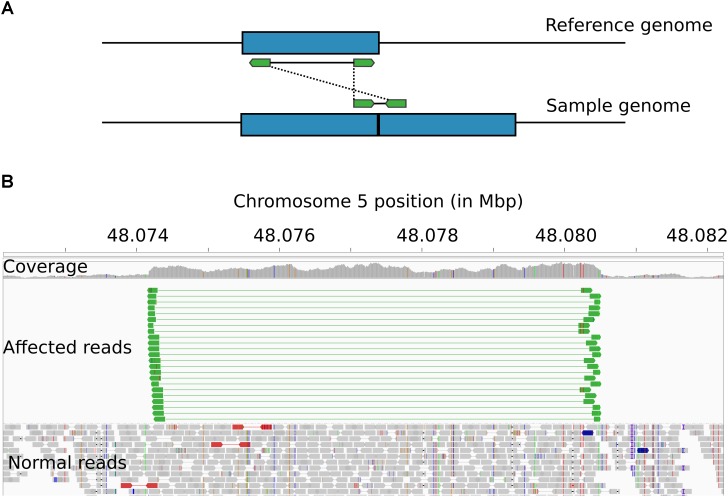
FIGURE 3.
Structural arrangement of the HMGA2-CNVR. (A) Schematic of the alignment of paired-end reads (in green) derived from an insert encompassing the boundaries of two repeats of a duplicated genomic segment (in blue). When the reads are aligned against the reference genome, the insert appears to be much larger than expected, usually spanning the entire length of the duplicated segment. Additionally, the tandem arrangement causes read pairs to be oriented toward the outer sides of the predicted insert. (B) IGV visualization of the HMGA2-CNVR in a whole-genome sequenced B. indicus animal showing several paired-end reads (in green) satisfying the pattern described above.