Fig. 1.
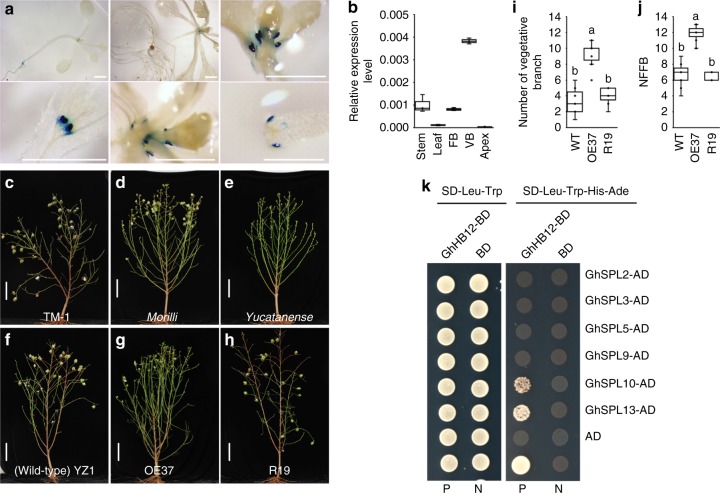
Identification of GhHB12. a Histochemical localization of GUS activity in proGhHB12:GUS Arabidopsis plants. Scale bars = 2 mm. b Detection of the expression of GhHB12 in stem, leaf, fruit branch bud (FB), vegetative branch bud (VB), and apex of domesticated upland cotton (Gossypium hirsutum L. YZ1) by qRT-PCR. The GhUBQ7 gene was used as the endogenous reference gene. The data represent the mean ± SD of three technical replicates. c–h Photographs of domesticated upland cotton TM-1 (c), YZ1 (f), OE37 (g, GhHB12 overexpression in the YZ1 background), and R19 (h, GhHB12 RNAi in the YZ1 background), ancestral upland cotton morilli (d) and yucatanense (e) plants. Scale bars = 20 cm. i and j Comparison of the number of vegetative branches (i) and the node of the first vegetative branch (NFFB) (j) among YZ1, OE37, and R19 plants. Error bars indicate the standard deviation of 12–15 plants, and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test). k Physical interaction of GhHB12 with GhSPL10 and GhSPL13 validated using the GAL-4-based Y2H assay. SD synthetic dropout, BD binding domain, AD activation domain, P positive control, N negative control