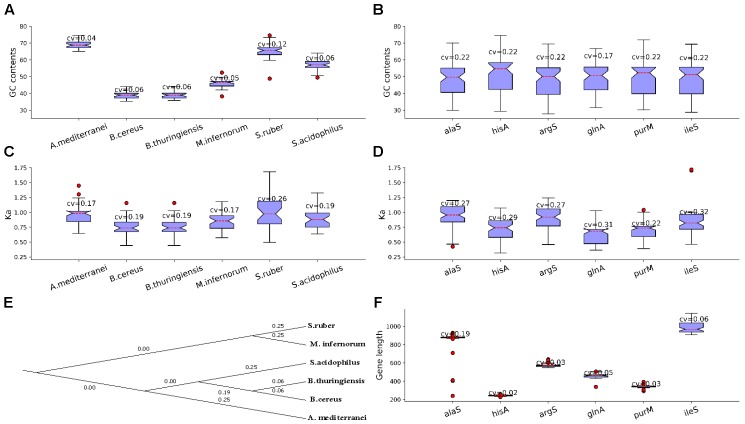
FIGURE 1.
Genes have biased GC contents and evolutionary rates in and among genomes. (A) The GC content of genes homologous to LUCA genes. The CV is short for Coefficient of Variation, which equals to standard deviation(x)/mean(x), while x is the list of GC contents of those genes in the LUCA list. Although homologous genes originate from the same gene and have identical GC contents in the ancestor, these genes tend to evolve to have preferred GC contents for the genomes. (B) Genes evolve to have variable GC content among genomes. By observing the GC content for evolved genes in genomes and among genomes, (A,B) show that GC content may play an important role for genome evolution. (C) The Ka of gene stes homologous to genes in the same genomes tend to be less discrete than that of the gene sets which contain homologous genes from different genomes (D), which means genes in one genome have similar evolutionary rates, while the genes among genomes evolve biasedly. (E) Shows the evolutionary relationship between the six presented organisms with K = 6, while using the MmarS2 as outgroup. (F) The six genes have different gene lengths. We chose these genes randomly, thus the evolutionary results may be not correlated with the gene length.