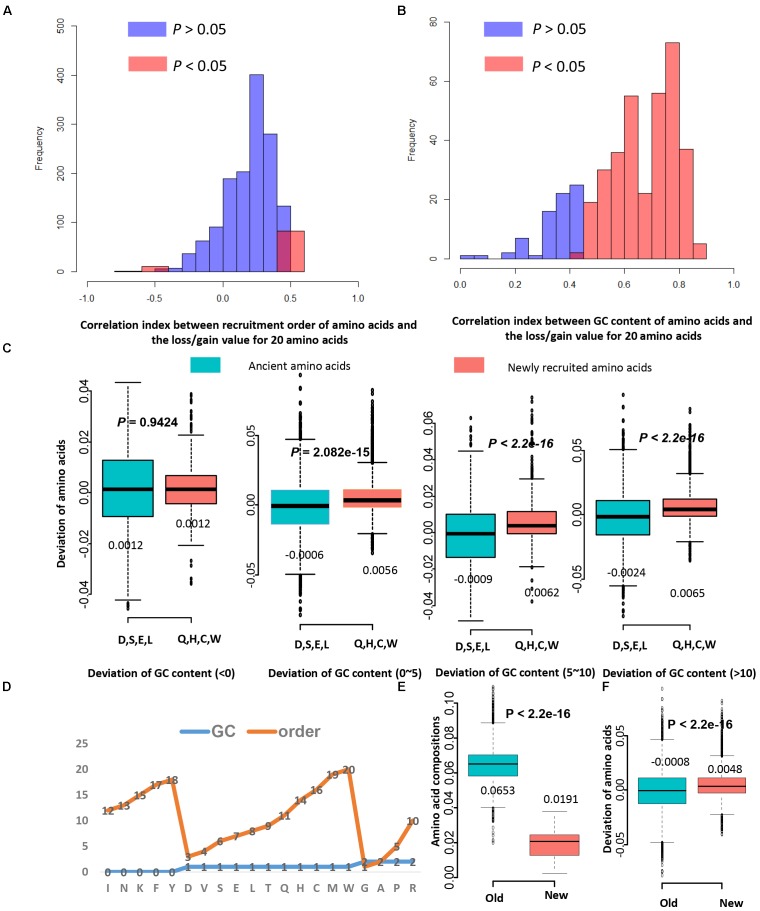
FIGURE 3.
The recruitment order of amino acids contributes less to the LUCA protein evolution than GC content. (A) The histograms for correlation indexes between recruitment order of amino acids and the loss/gain value for 20 amino acids. Only in 19.33% proteins (1501 records from 65 genomes) the deviation of amino acids correlates with the amino acid recruitment order. (B) The histograms for correlation indexes between GC content of amino acids and the loss/gain value for 20 amino acids. In 62.73% proteins the GC content significantly correlates with the deviation of amino acids. (C) The boxplots of values of deviation of amino acids for ancient amino acids and newly recruited amino acids. Four ancient amino acids D, S, E, L (Asp, Ser, Glu, Leu) tend to be lost during evolution in proteins homologous to LUCA proteins, while four newly recruited amino acids Q, H, C, W (Gln, His, Cys, Trp) tend be gained. (D) The order and GC content features for 20 standard amino acids. The GC-rich amino acids are with values of 2, and the AT-rich amino acids are with values of 0. The early recruited amino acids are from 1 to 10, and the newly recruited amino acids are from 11 to 20. (E) The amino acid compositions for old amino acids (D, V, S, E, L, T) are significantly higher than that of new amino acids (Q, H, C, W) for 2774 bacteria and archaea genomes. (F) The deviation of amino acids for old amino acids (D, V, S, E, L, T) are significantly lower than that of new amino acids (Q, H, C, W) for 1501 proteins. These proteins were used in panels (A,B). Although new amino acids have lower amino acid levels, they may have higher deviations. Thus, the low levels of new amino acid compositions may not cause the lower chance of amino acid replacement occurrence.