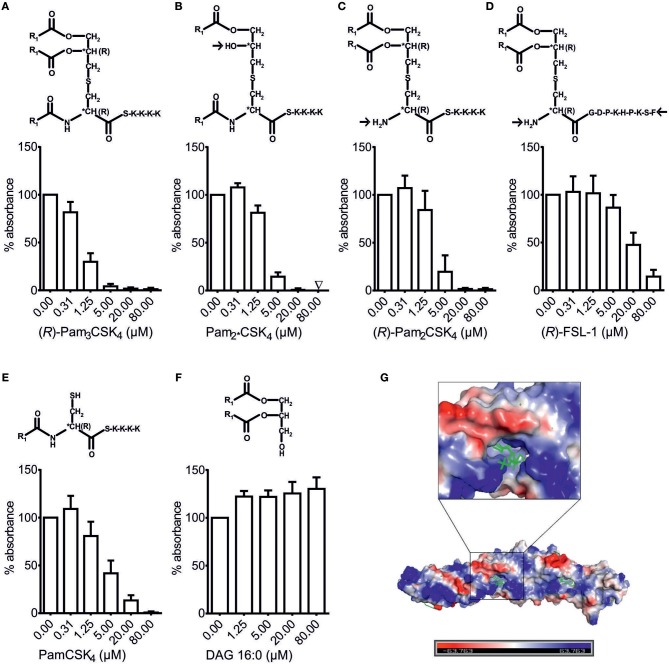
Figure 4.
Structural requirements for binding of bLPs to BPI. BPI binding assay (A–F). Lipopeptides with modified structure as well as DAG were tested for their potential to inhibit binding of rBPI to LPS biotin-coated wells (A–F). Absorbance measured at 450 nm for wells with rBPI alone was set to 100% to ensure comparability between the different ligands. All results are shown as means ± SD of at least four biological replicates. R1 = CH3(CH2)14. Electrostatic charge surface of the BPI structure (G). N-terminal lipid-binding pocket surrounded by differently charged areas is depicted with phosphatidylcholine as ligand.