Figure EV2. CDKLs 1–4 cannot phosphorylate MAP1S or CEP131 in cells.
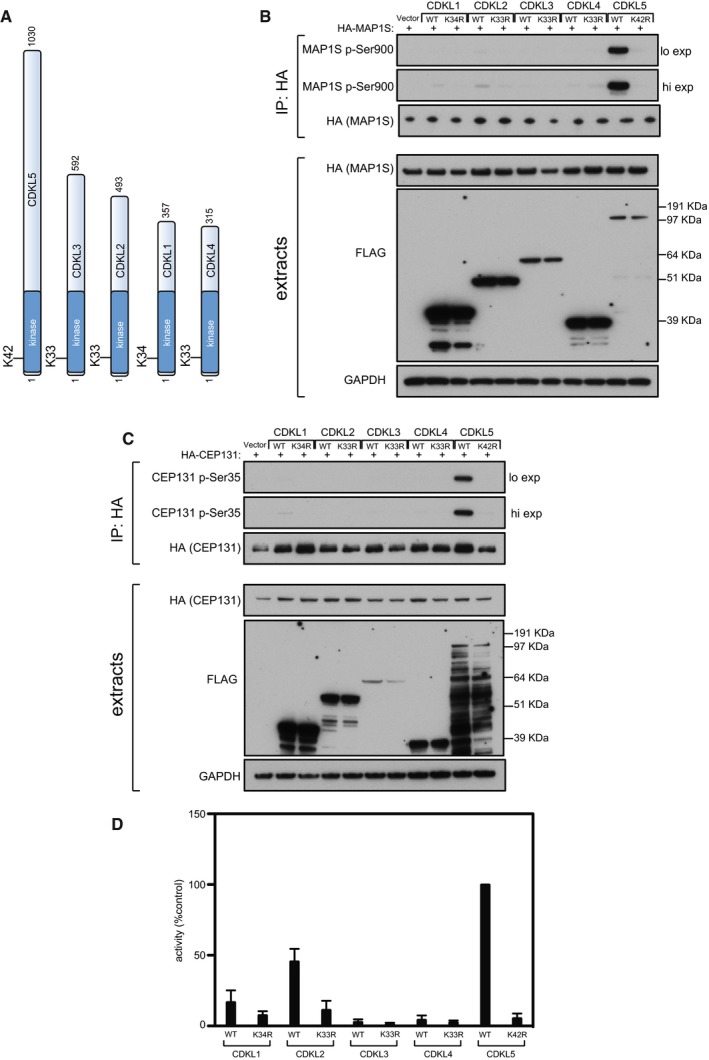
- Schematic representation of CDKLs 1–5. The kinase catalytic domain is highlighted in dark blue. Amino acid numbers at the N‐ and C‐termini are indicated; the positions of conserved residues in the ATP binding sites that were mutated to render these protein “kinase‐dead” are also indicated.
- HEK293 cells were co‐transfected with C‐terminally tagged FLAG‐tagged CDKLs 1,2,3,4 or 5 (wild type “WT” or the relevant kinase‐dead mutant) and HA‐MAP1S. Anti‐HA precipitates were subjected to Western blotting with the antibodies indicated. “Hi” higher exposure; “lo” lower exposure. The input extracts were also subjected to immunoblotting (lower panels). Three independent experiments were done, and one representative experiment is shown.
- Same as (B) except that HEK293 cells were co‐transfected with C‐terminally tagged FLAG‐tagged CDKLs 1,2,3,4 or 5 (wild type “WT” or the relevant kinase‐dead mutant) and HA‐CEP131. Three independent experiments were done, and one representative experiment is shown.
- HEK293 cells were transfected with C‐terminally tagged FLAG‐tagged CDKLs 1,2,3,4 or 5 (wild type “WT” or the relevant kinase‐dead mutant). Anti‐FLAG precipitates were incubated with the MAP1S S900 synthetic peptide in the presence of [γ‐32P]‐labelled ATP‐Mg2+, and peptide phosphorylation was measured by Cerenkov counting. Data are represented as mean ± SEM from three independent experiments.
Source data are available online for this figure.
