Figure 4. CDKL5 sequence determinants favouring phosphorylation by CDKL5.
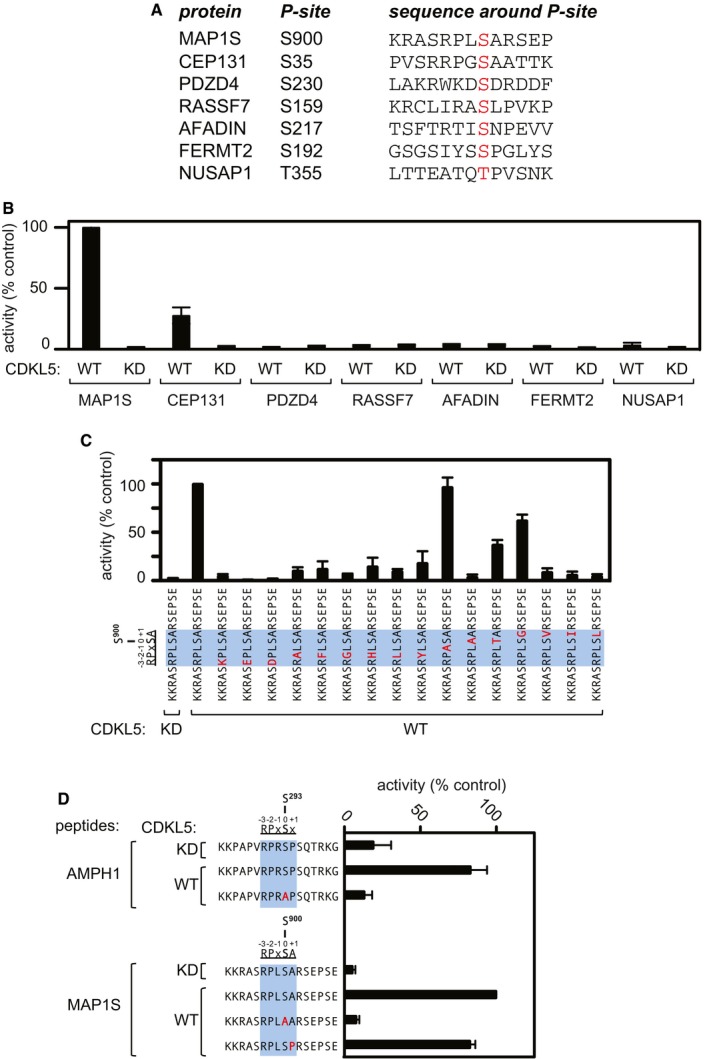
- Sequence of synthetic peptides surrounding the sites of phosphorylation in putative CDKL5 substrates that were identified in the phosphoproteomic screening. The amino acid number of the phosphorylated residue in each peptide (highlighted in red) is listed.
- Peptide kinase assays to investigate CDKL5 sequence specificity. Anti‐FLAG precipitates from HEK293 cells transiently expressing FLAG‐tagged CDKL5 (wild type “WT” or a K42R kinase‐dead “KD” mutant) were incubated with the synthetic peptides from the proteins indicated (sequences shown in A) in the presence of [γ‐32P]‐labelled ATP‐Mg2+, and peptide phosphorylation was measured by Cerenkov counting.
- Same as (B), except that the peptides used were designed specifically to investigate the effect of amino acid substitutions at R897, P898, L899 and A901 on the phosphorylation of MAP1S Ser900. The RPXSA motif is shaded in blue, and amino acid substitutions compared with the wild‐type MAP1S Ser900 peptide are shown in red.
- Same as (C), except that phosphorylation of the indicated peptides from AMPH1 and MAP1S was compared.
