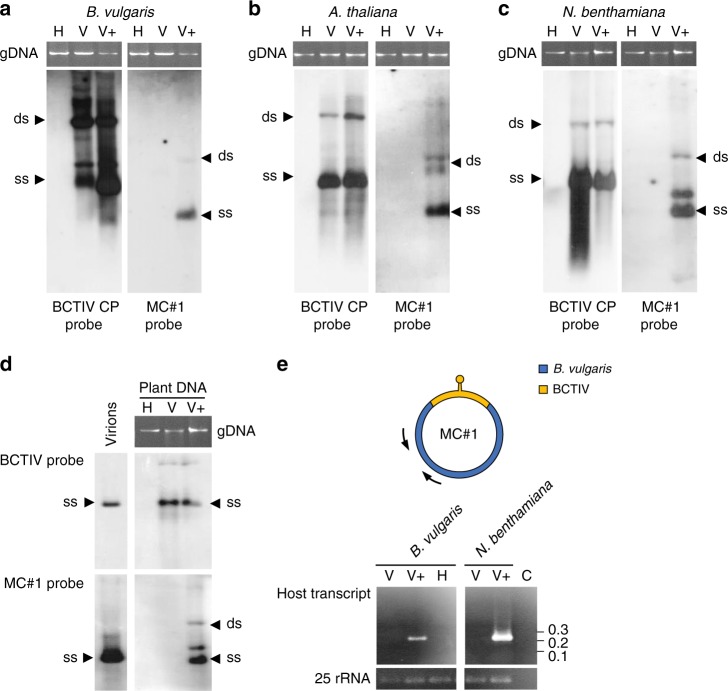
Fig. 3.
MC#1 replication and transcription when transfected to various hosts. Southern blots of DNA extracted from B. vulgaris (a), A. thaliana (b) and N. benthamiana (c), mock-inoculated healthy plants (H); plants agroinfected with BCTIV (V) or with both BCTIV and MC#1 (V+). Leaves were collected at 4 weeks post inoculation (wpi) and hybridized with BCTIV- or MC#1-specific probes (see Fig. 1b). Bands of single and double stranded DNAs are indicated as “ss” and “ds”, respectively. Plant genomic DNA (gDNA) is shown above the blots as a loading control. Source data are provided as Supplementary Data 5. d Southern blot of DNA extracted from purified BCTIV virions and from the N. benthamiana plants used for purification of BCTIV particles, hybridized with the BCTIV- and the MC#1-specific probes (see Fig. 1b). “ss”, “ds” and gDNA as in a–c. Source data are provided as Supplementary Data 5. e Transcription of the beet-derived DNA sequence of MC#1 in B. vulgaris and N. benthamiana plants inoculated with BCTIV and MC#1 (V+) detected by RT-PCR with primers positioned as indicated by arrows in the upper part of the panel. Lanes are marked H, V and V+ as in a–d, whereas C is a RT-PCR-negative reaction control. Source data are provided as Supplementary Data 5