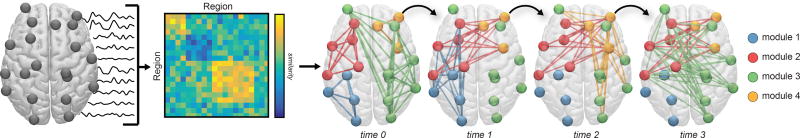
Fig. 1. Brain graphs and communities within them.
One can construct a brain graph in several ways, and subsequently study its modular architecture using community detection techniques developed for graphs. Here, we illustrate an example pipeline in which we use non-invasive neuroimaging in humans to obtain regional timeseries of continuous neural activity (Left). Next, we define a weighted undirected graph and represent that graph in an adjacency matrix, each element of which provides an estimate of the statistical similarity between the time series of region i and the time series of region j (Middle). Finally, we apply community detection techniques to the brain graph to identify modules. Here, a module is composed of nodes (regions) that are more densely interconnected with one another than expected in some appropriate random network null model. If we have temporally extended data, we can also consider defining a temporal graph, and using dynamic community detection techniques to study the temporal evolution of modules and their relation to cognition (Right). In this review, we discuss considerations, methods, statistics, and interpretations relevant to this process.