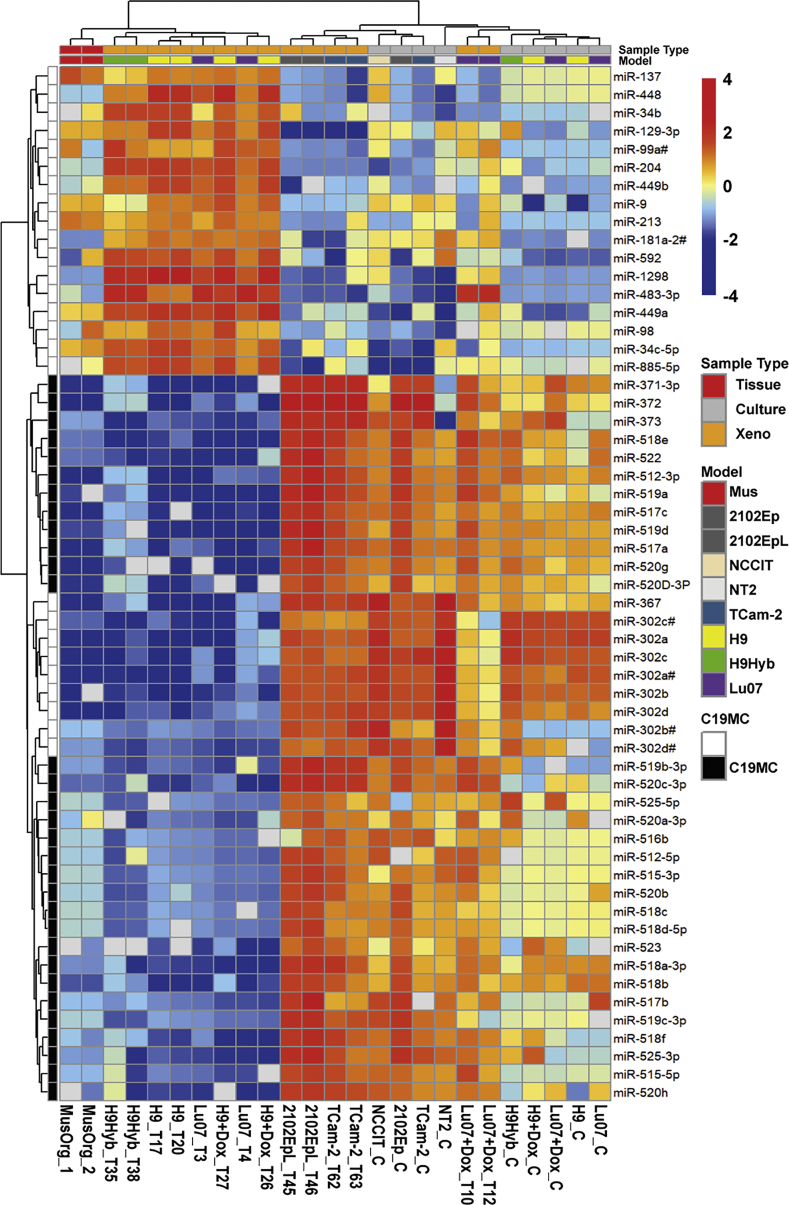
Figure 4.
miRNA Expression Profile Comparison between hPSCs and hGCT-Derived Cell Lines and Respective Xenografts
The miRNA expression of hPSC cell lines (Lu07 + Dox_C, Lu07_C, H9_C, H9 + Dox_C, and H9Hyb_C), hGCT-derived cell lines (NCCIT_C, 2102Ep_C, NT2_C, and TCam-2_C) and xenografts derived from the respective hPSC (Lu07 + Dox_T, Lu07_T, H9_T, H9 + Dox_T, and H9Hyb_T) and hGCT (2102EpL_T and TCam-2_T) cell lines was determined and compared. miRNA profiles of NSG mouse organs were included as internal control (MusOrg). The heatmap shows 57 of the 768 examined miRNAs of which the levels were detectable in the majority of the samples (average Cq < 36). The color key shows the relative levels of the miRNAs. Color codes for specific Sample Type and Model are indicated on the right. miRNAs located in the primate specific chromosome 19 miRNA cluster (C19MC) are indicated. Raw data are available in Table S4.