Fig. 1.
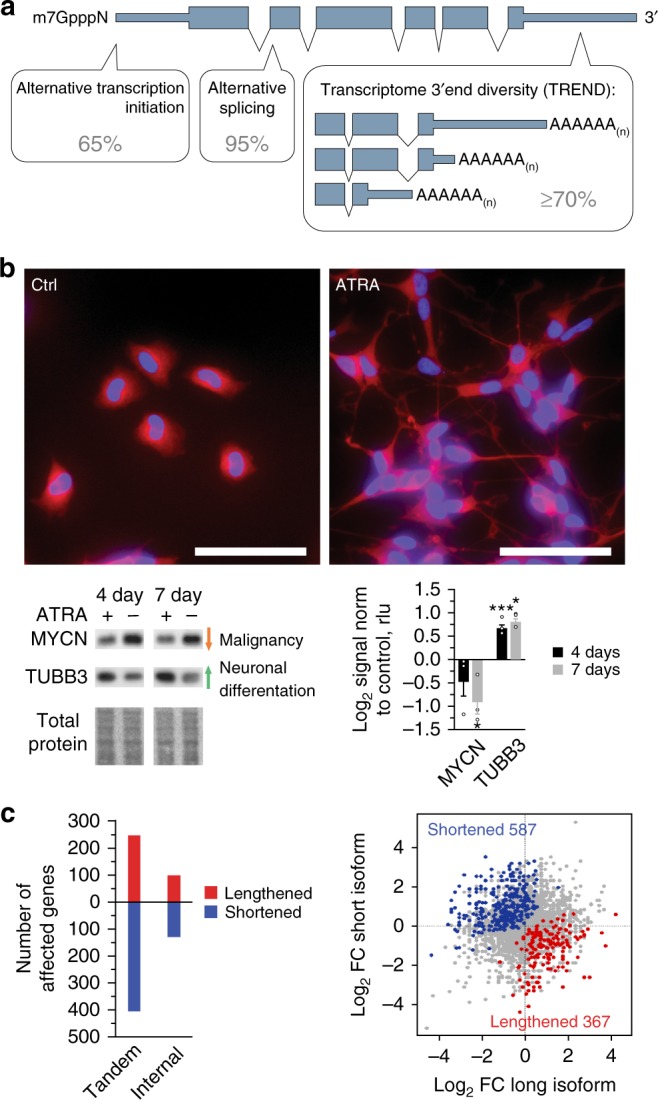
Pervasive alteration of transcriptome 3′end diversification (TREND) in childhood neuroblastoma. a The genome complexity is considerably expanded by co- and post-transcriptional processes. Various mechanisms including alternative polyadenylation (APA) can result in transcriptome 3′end diversification (TREND) affecting the coding sequence and/or 3′untranslated regions (UTR). More than 70% of all genes expressed have alternative 3′ ends, thus shaping the transcriptome complexity to a similar extent as alternative splicing (95%) or alternative transcription initiation (65%). b BE(2)-C neuroblastoma model for all-trans retinoic acid (ATRA) induced neurodifferentiation (scale bar 100 µm). Western blot of molecular markers reflecting neuronal differentiation in response to ATRA treatment (mean ± s.e.m. for three replicates, one-sided t-test p-value, *p < 0.05, ***p < 0.001; cells are stained with an antibody directed against TUBB3 (red), nuclei are stained with DAPI (blue)). c Differentiation results in a widespread TREND regulation leading to the expression of lengthened (red) and shortened (blue) transcript isoforms, mainly affecting the 3′UTR (‘tandem’; also depicted in the scatter plot on the right) compared to relatively fewer events affecting the open reading frame (‘internal’; FC fold change)
