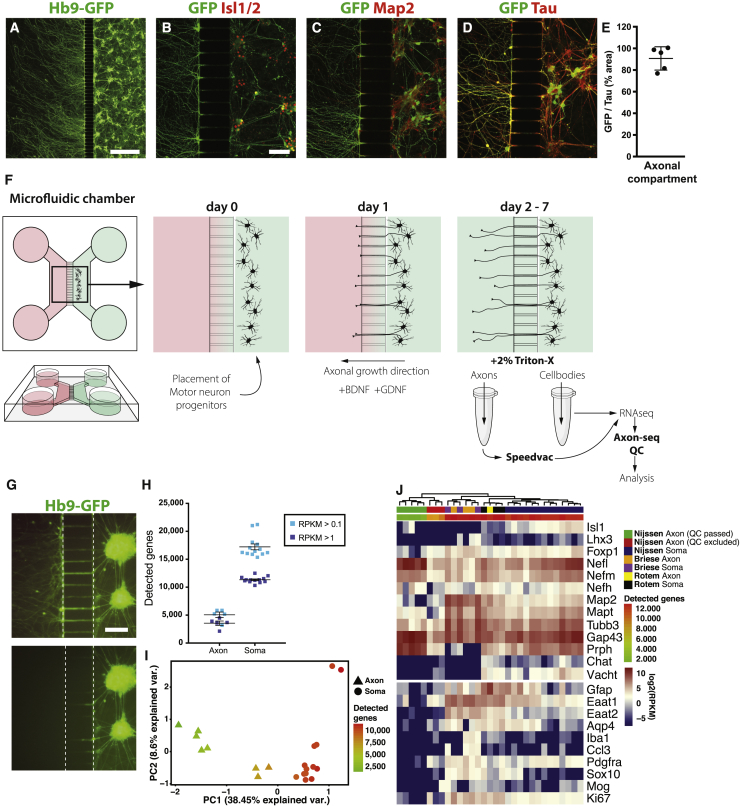
Figure 1.
Axon-Seq of Motor Axons in Single Microfluidic Devices
(A and B) Anti-GFP immunofluorescence visualizes motor axons crossing the microgrooves and extending into the axonal compartment. MN somas are visualized with (A) Hb9-eGFP and (B) Isl1/2 staining.
(C and D) Presence of Tau (Mapt) (D) but very little Map2 (C) indicates that mainly axons cross over into the other compartment.
(E) Quantification of Hb9::eGFP+ area over Tau+ area reveals that the majority of crossing axons are motor axons (90.75% ± 4.8%). Data are represented as mean ± SEM.
(F) Overview of the methodology used. MNs are cultured in one compartment of the microfluidic device and axons are recruited across microgrooves using a gradient of trophic factors. Compartments are separately lysed and cDNA libraries prepared for RNA sequencing.
(G) Hb9-eGFP expression reveals that the somatic chamber is not affected by lysis of the axons.
(H) The axonal fractions contain around 5,000 unique transcripts, while MN somas contain >15,000 (mean ± SEM).
(I) PCA based on all expressed genes showing “contaminated” axon samples with an intermediate number of detected genes that cluster away from the clean axon samples and toward the somatodendritic samples.
(J) Expression of selected marker genes in soma and axon samples of this study and data from studies by Rotem et al. (2017) and Briese et al. (2016). Our clean axon samples separate from the rest, while axon and soma samples from previously published studies intermingled, indicating cross-contamination.
Scale bars: (A) 500 μm. (B) 100 μm; scale bar in (B) also applies to (C) and (D). (G) 100 μm.