Figure 2.
Axons Have a Transcriptome Distinct from that of Somas
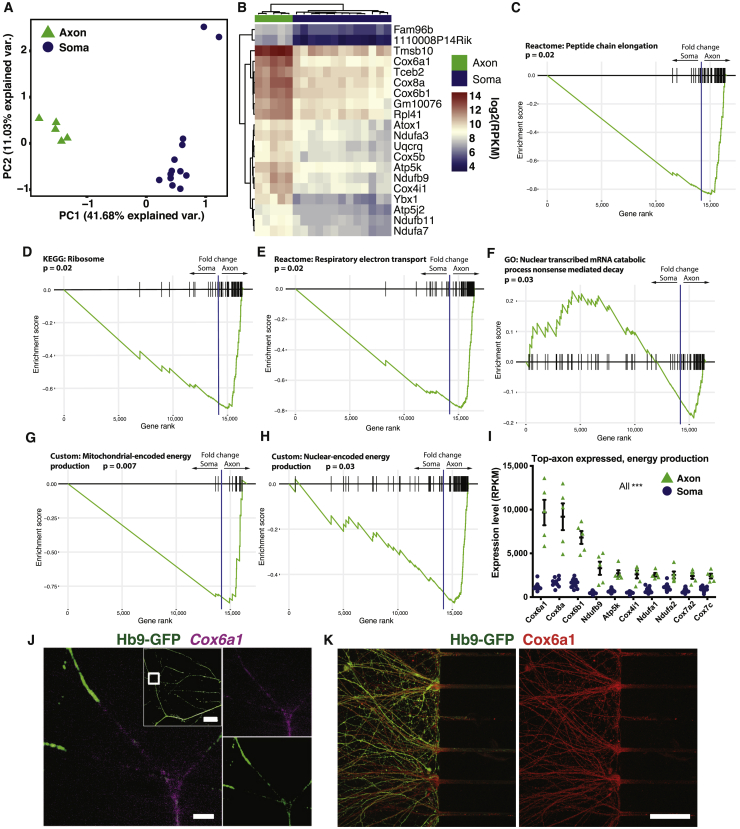
(A) PCA of all expressed genes reveals a clear separation of axons and somas.
(B) Top 20 genes determining PC1-negative loading. All of them are strongly enriched in axons compared with somas.
(C–H) Gene set enrichment analysis (GSEA) using fold changes from the DESeq2 differential expression output ranked list between axons and somas, without a p value cutoff. GSEA uncovers pathways related to (C and D) local translation, (E) energy production, and (F) nonsense-mediated decay. (G and H) Subdividing the oxidative energy production class into (G) mitochondrial- and (H) nuclear-encoded transcripts reveals an axonal enrichment for both. Differential expression was performed on the five axon samples that passed QC and 13 soma samples, as visualized in (A) and (B).
(I) Expression levels of the highest expressed transcripts in axons related to oxidative energy production. All of the genes plotted are significantly enriched in the axonal compared with the somatodendritic compartment (∗∗∗p < 0.001). p values are derived from the DESeq2 differential expression and are adjusted for multiple testing. Data are represented as mean ± SEM.
(J) RNAscope and (K) immunocytochemistry of Cox6a1, the most abundant mRNA in axons involved in energy production. Scale bars: (J) 2 μm, lower magnification inset, 20 μm. (K) 50 μm.