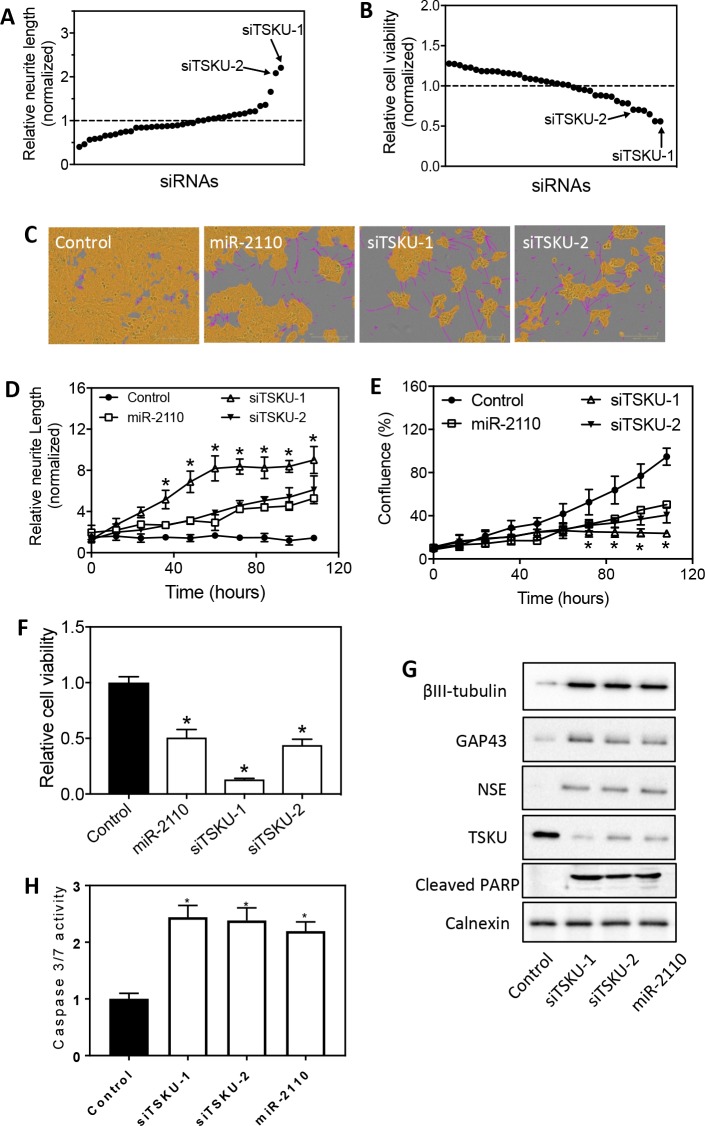
Fig 2. Identification of TSKU as miR-2110 target that regulates BE(2)-C cell differentiation.
(A-B) HCS of siRNAs against the 21 predicted target genes of miR-2110 identifies siRNAs that induce neurite outgrowth and reduce cell viability in BE(2)-C cells. Cells were transfected with each siRNA (25 nM) in triplicate and cultured for 4 days. Neurite outgrowth and cell viability was measured as above. A, Scatter plot of normalized neurite lengths, sorted in ascending order, associated with individual siRNAs. B, Scatter plot of normalized cell viabilities, sorted in descending order, associated with individual siRNAs. Shown are the mean values of the normalized neurite lengths and cell viabilities derived from three independent experiments. Dotted lines are the means of the library. (C-F) Effect of siTSKUs on neurite outgrowth and cell viability was validated in separate experiments using re-ordered siTSKUs. Cells were transfected with the indicated oligos (25 nM) and neurite outgrowth and cell confluence were measured every 12 hours for 4 days. Cell viability was measured at the end-point of the experiment. C, Representative cell images analyzed to define neurites and cell body areas after 4 days of treatment. (D-E), Time-dependent neurite outgrowth (D) and cell confluence change (E) under each treatment. *, p < 0.05 for all three treatments (miR-2110, siTSKU-1 and siTSKU-2) compared to the control at the respective time points. F, Cell viability associated with each treatment. *, p < 0.05 compared to control. (G) Effect of siTSKUs and miR-2110 mimic on protein levels of cleaved PARP and cell differentiation markers. Cells were transfected and protein levels were measured as above. (H) Effect of siTSKUs and miR-2110 mimic on caspase 3/7 activity. *, p < 0.05 compared to control.