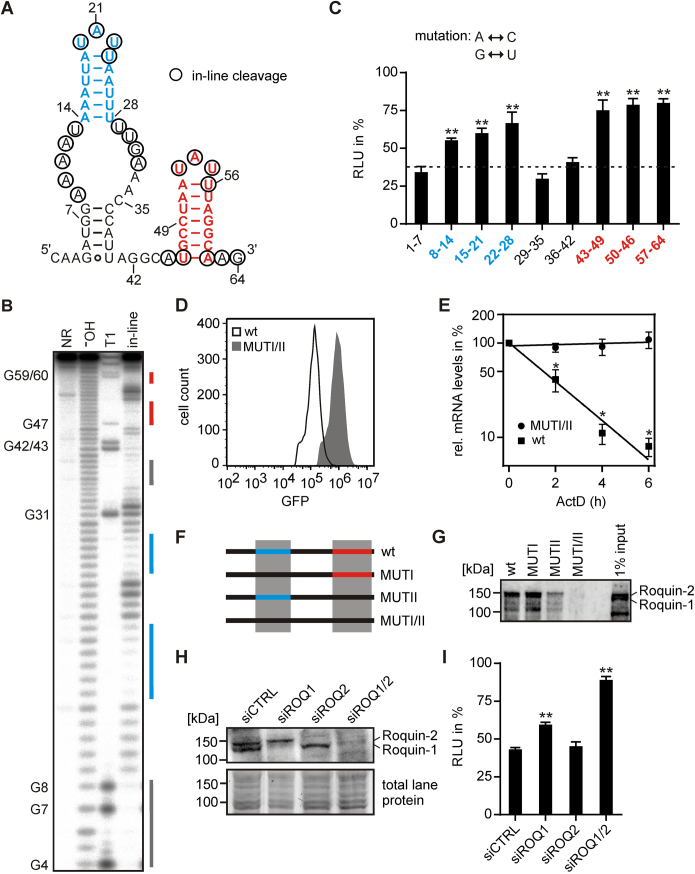
Figure 2.
The UCP3 wt element folds into two hairpins and reduces mRNA half-life by interaction with Roquin. (A) Predicted lowest free energy secondary structure of the UCP3 wt element by RNAstructure. Nucleotides detected by in-line probing [shown in (B)] are circled. (B) In-line probing analysis of UCP3 wt RNA. The RNA was loaded directly (NR, no reaction), subjected to cleavage by RNase T1 or alkaline hydrolysis (¯OH), or incubated for 40 h at room temperature and pH 8.3 (in-line) prior to Urea PAGE. Paired regions are indicated by identically colored lines. (C) Luciferase activity of UCP3 mutants for the identification of motifs essential for gene regulation. Adenine was mutated to cytosine, guanine to uracil and vice versa. Numbers indicate mutated nucleotide positions. Firefly luciferase activity was normalized to Renilla luciferase as internal transfection control. Values are normalized to an empty vector control, without UCP3 3′UTR sequences. Luciferase activity of the UCP3 wt element is indicated as dashed line. n = 3. (D) GFP fluorescence of UCP3 wt and double mutant (MUTI/II). GFP-UCP3-fusion constructs were stably integrated into the genome of HeLa cells. GFP fluorescence was measured by flow cytometry. (E) Half-life of GFP mRNAs containing the UCP3 wt element (wt) or double mutant (MUTI/II). HeLa cells stably expressing one of the two constructs were treated with 5 μg/μl actinomycin D (ActD). Thereafter, total RNA was isolated at 2 h intervals and GFP mRNA levels quantified by RT-qPCR. GFP values are normalized to the housekeeping gene RPLP0. n = 3. (F) Overview of UCP3 constructs used for RNA affinity purification. (G) Analysis of Roquin binding to the UCP3 constructs shown in (F). For RNA affinity purification HEK293 whole cell lysates were incubated with the different UCP3 RNAs. Roquin-1 and Roquin-2 were visualized by western blot using anti-Roquin antibody. n = 2. (H) Western blot of Roquin-1 and Roquin-2 after siRNA-mediated knockdown. Anti-Roquin was used to verify the respective knockdown. Total lane protein is shown as loading control. n = 3. (I) Luciferase activity of the UCP3 wt element after siRNA-mediated knockdown of Roquin proteins. Firefly luciferase activity was normalized to Renilla luciferase as internal transfection control. Values are normalized to an empty vector control, without UCP3 3′UTR sequences. n = 3. (**) P-value < 0.01. (*) P-value < 0.05.