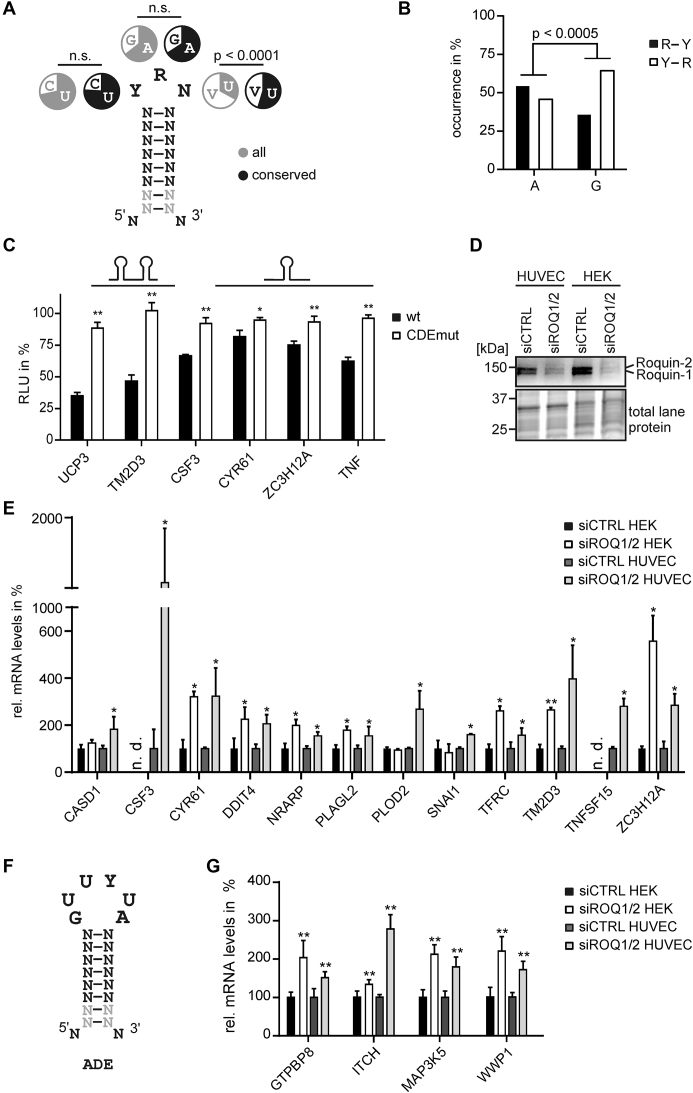
Figure 5.
Identification of new Roquin targets. (A) Consensus used for bioinformatic prediction of new CDE structures by RNAMotif. Nucleotide occurrences in the triloop are shown for all and conserved elements. V = A, G, C. P-value < 0.0001 (Pearson’s chi-square test). (B) Co-occurrence of A or G at position 2 in the triloop with R-Y or Y-R closing base pairs. R = A, G and Y = C, U. P-value < 0.0005 (Pearson’s chi-square test). (C) Luciferase activity of different CDE-encoding 3′UTR regions and mutants. n = 3. CDEs and mutations are shown in Supplementary Figure S13. Firefly luciferase activity was normalized to Renilla luciferase as internal transfection control. Values are normalized to an empty vector control, without UCP3 3′UTR sequences. Influence of predicted CDE-like elements on luciferase activity in HEK293 cells. n = 3. (D) Western blot of Roquin-1 and Roquin-2 after siRNA-mediated knockdown in HEK293 cells and HUVECs. Anti-Roquin was used to verify the respective knockdown. Total lane protein is shown as loading control. n = 3. (E) RT-qPCR quantification of new Roquin target genes encoding CDEs after siRNA-mediated knockdown of Roquin-1 and Roquin-2 in HEK293 cells and HUVECs. Values are normalized to the housekeeping gene RPLP0. n = 4. (F) Consensus used for bioinformatic prediction of new ADE structures by RNAMotif. (G) RT-qPCR quantification of new Roquin target genes encoding ADEs after siRNA-mediated knockdown of Roquin-1 and Roquin-2 in HEK293 cells and HUVECs. Values are normalized to the housekeeping gene RPLP0. n = 4. (**) P-value < 0.01. (*) P-value < 0.05. n.d. = not detected.