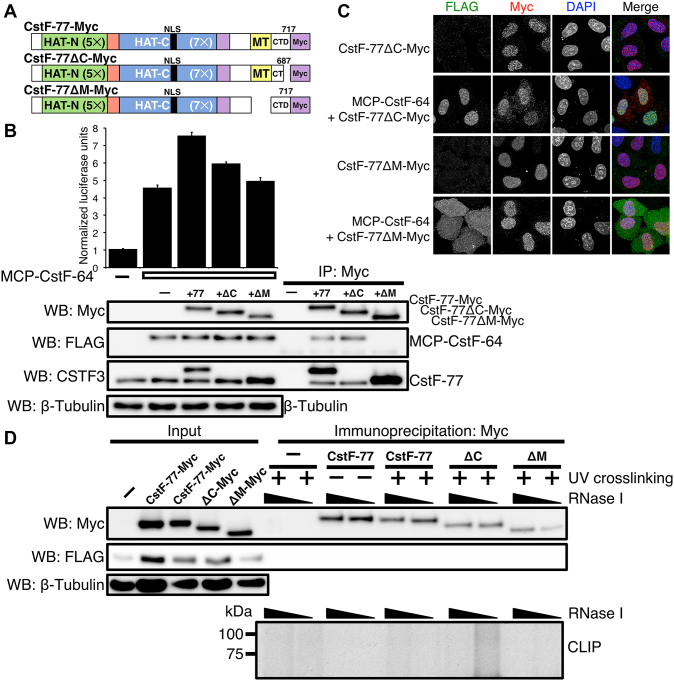
Figure 6.
The CTD of CstF-77 is involved in modulating the binding affinity of the RRM of CstF-64. (A) Illustration of CstF-77-Myc deletion mutants: the last 30 amino acids of the carboxy terminal domain (amino acids deleted Δ688–717, CstF-77ΔC-Myc), and MT Δ607–664, CstF-77ΔM-Myc). (B) SLAP of the wild-type and CstF-77-Myc deletion mutants. Western blots to verify the expression of the proteins: antibodies against Myc tag to detect CstF-77-Myc construct, FLAG to detect MCP-CstF-64 and against CSTF3 to detect endogenous and exogenous CstF-77 protein. Note that the antibody against CSTF3 is raised against a peptide within the last 30 amino acids of the CstF-77 and therefore does not recognize the CstF-77ΔC-Myc construct. (C) Immunohistochemistry staining of the CstF-77-Myc deletion mutants (red) alone or co-transfected with MCP-CstF-64 (green). DNA in the nucleus is counterstained with DAPI. (D) CLIP (bottom) of the CstF-77-Myc deletion mutants. UV-crosslinking or the lack of it is indicated. The increasing amount of RNase I is also indicated. Amount of the proteins in the input is shown for MCP-CstF-64 (WB: FLAG), wild-type CstF-77-Myc and deletion mutants (WB: Myc). β-tubulin was used as a loading control. Note that in the conditions of CLIP using RIPA buffer, MCP-CstF-64 does not co-immunoprecipitated with CstF-77-Myc.