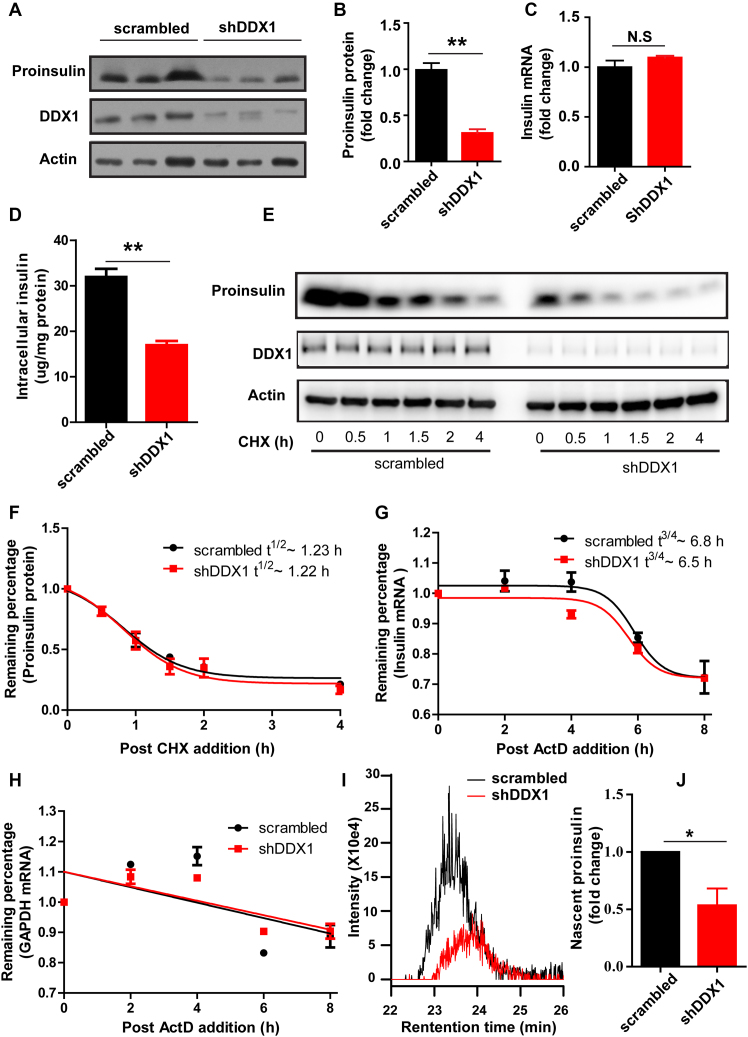
Figure 4.
Knockdown of DDX1 affects insulin translation. (A–D) Western blotting was used to determine proinsulin expression in INS-1 cells stably expressing shRNA against DDX1 (shDDX1) and in INS-1 cells stably expressing scrambled shRNA (scrambled) as a control (A). The fold change in proinsulin expression was calculated by normalization to actin expression (B). RT-qPCR was used to determine the insulin mRNA levels (C). ELISA was used to detect the intracellular insulin content (D). (E, F) To assess whether the knockdown of DDX1 affected the stability of proinsulin, scrambled and shDDX1 cells were treated with cycloheximide to block nascent protein synthesis, and the stability of the proinsulin protein was assessed by western blotting (E). The fold change in proinsulin expression was calculated by normalization to actin expression (F). (G, H) To assess whether the knockdown of DDX1 affected the stability of insulin mRNA, scrambled and shDDX1 cells were treated with the transcription inhibitor actinomycin D, and RT-qPCR was used to determine the remaining insulin (G) and Gapdh (H) mRNA levels. (I, J) LC/MS was used to detect nascent proinsulin biosynthesis in scrambled and shDDX1 stable cell lines (I). The nascent proinsulin level was normalized to the total protein level (J). The results are shown as the means ± SEMs (n = 3). Significance was determined by a two-tailed unpaired t-test; *P < 0.05; **P < 0.01; N.S, no significance.