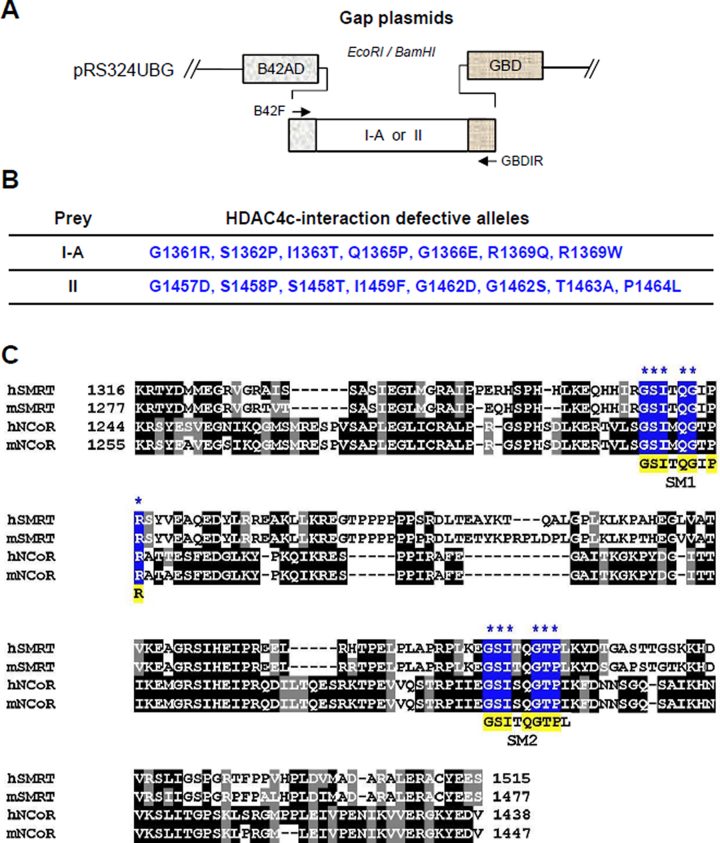
Figure 1.
OPTHiS screening identifies two SRD3c conserved motifs required for HDAC4c binding. (A) Schematic diagram of the prey constructs used for OPTHiS screening. Randomly mutagenized I-A or II fragments were inserted between the B42AD and the GBD regions of gap plasmid to construct the prey mutant library for OPTHiS screening. (B) Positions and amino acid changes of isolated I-A or II mutant alleles defective for HDAC4c binding. (C) Multiple sequence alignments of RD3c regions of SMRT and NCoR proteins from H. sapiens (h), M. musculus (m). Sequence alignment was performed and presented using the ClustalW and Boxshade programs, respectively. Positions and amino acids of the isolated mutations are marked with stars and shaded in blue. Conserved amino acids within SM1 and SM2 motifs are shaded in yellow.