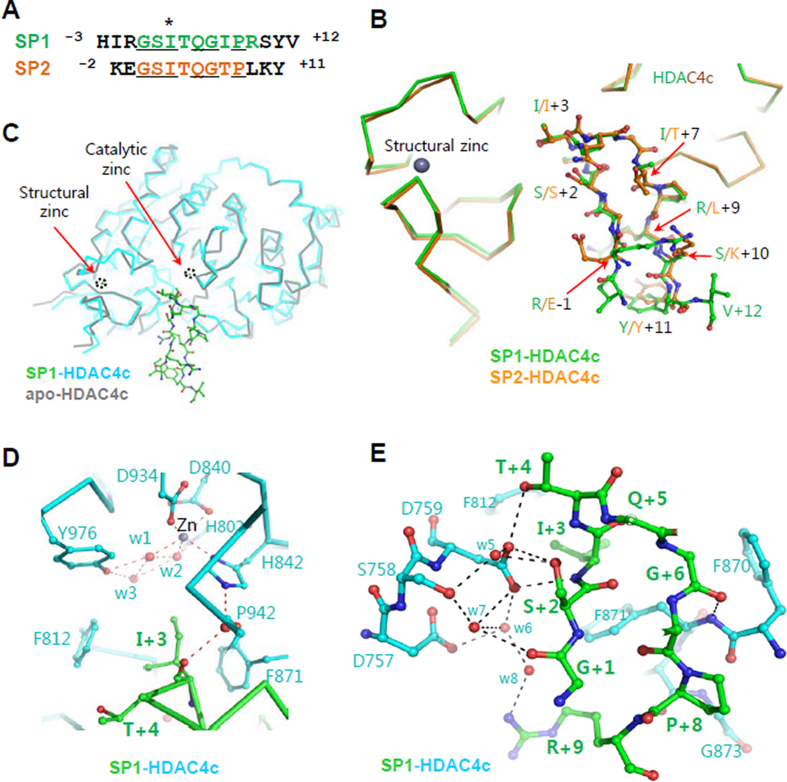
Figure 3.
HDAC4c structures in complex with SMRT peptides. (A) Sequence alignment of SP1 and SP2 peptides derived from two SMRT consensus motifs. The I+3 residues (I1363 and I1459) sandwiched between the two conserved phenylalanine residues (F812 and F871) of HDAC4c are indicated by an asterisk. The strictly conserved residues are underlined. (B) Superposed structures of HDAC4c in complex with two peptides. HDAC4c and bound peptides are shown in different colors. Peptide side-chains are displayed with the ball-and-stick models. The structural zinc ion is drawn as a black sphere. (C) Superposed structures of closed conformation (gray) and the peptide complex (cyan-green). The catalytic and structural zinc ions are drawn as spheres and indicated with black-dotted circles. (D) A close-up view of the catalytic zinc ion-binding site in the peptide-complex structure. The zinc ion and water molecules are drawn as gray and red spheres, respectively. The residues of HDAC4c (cyan) and bound SP1 peptide (green) are displayed with the ball-and-stick models. The interactions among atoms are displayed by red-dotted lines. (E) Interaction of SP1 peptide with HDAC4c at the cleft to the catalytic site. Water molecules are displayed with red spheres and interacting residues are presented with the ball-and-stick models. Polar interactions among atoms are indicated with black-dotted lines.