Abstract
LAGLIDADG homing endonucleases (meganucleases) are site-specific mobile endonucleases that can be adapted for genome-editing applications. However, one problem when reprogramming meganucleases on non-native substrates is indirect readout of DNA shape and flexibility at the central 4 bases where cleavage occurs. To understand how the meganuclease active site regulates DNA cleavage, we used functional selections and deep sequencing to profile the fitness landscape of 1600 I-LtrI and I-OnuI active site variants individually challenged with 67 substrates with central 4 base substitutions. The wild-type active site was not optimal for cleavage on many substrates, including the native I-LtrI and I-OnuI targets. Novel combinations of active site residues not observed in known meganucleases supported activity on substrates poorly cleaved by the wild-type enzymes. Strikingly, combinations of E or D substitutions in the two metal-binding residues greatly influenced cleavage activity, and E184D variants had a broadened cleavage profile. Analyses of I-LtrI E184D and the wild-type proteins co-crystallized with the non-cognate AACC central 4 sequence revealed structural differences that correlated with kinetic constants for cleavage of individual DNA strands. Optimizing meganuclease active sites to enhance cleavage of non-native central 4 target sites is a straightforward addition to engineering workflows that will expand genome-editing applications.
INTRODUCTION
Homing endonucleases are site-specific DNA endonucleases that are typically encoded within self-splicing introns or inteins and that function as mobile elements (1). Six families of homing endonuclease have been identified to date (2), including the LAGLIDADG endonuclease family that is widespread in microbial and organellar genomes. LAGLIDADG enzymes, also called meganucleases, occur as homodimers or gene-fused single-chain monomers. In either case, the class-defining LAGLIDADG amino acid motif forms a parallel α-helical interface with the catalytic metal-binding residue (underlined and bold LAGLIDADG) positioned at the bottom of each helix in close proximity to the DNA cleavage sites (Figure 1) (3–5). Meganucleases are of evolutionary interest because their mobility impacts genome structure and function, and of biotechnological interest because they represent one of the few nuclease platforms where specificity can be re-programmed (6–12).
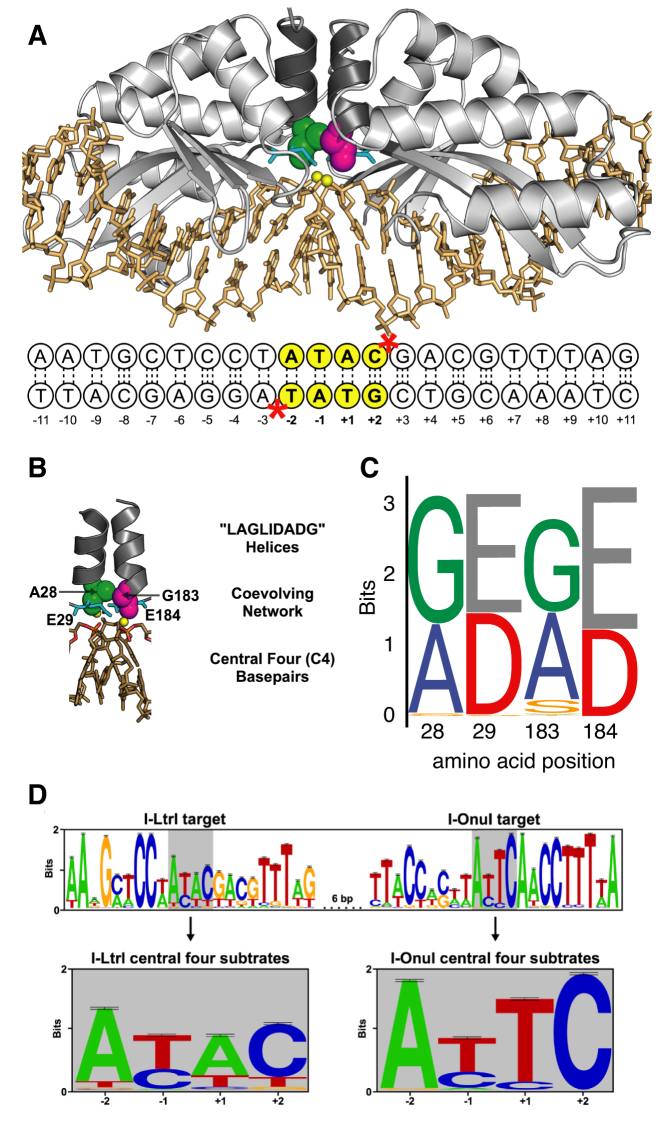
Figure 1.
Overview of I-LtrI structure and target site preference. (A) Structural model of I-LtrI bound in a pre-cleavage complex to its native recognition site (modified from PDB: 3R7P). Shown are I-LtrI secondary structure elements (light gray), the LAGLIDADG helices (dark gray), A28 (green spheres), G183 (magenta spheres), E29 and E184 (cyan sticks), calcium ions (yellow spheres) and the cognate DNA sequence (gold sticks) with each position numbered. Dotted lines represent hydrogen bonds. The central 4 bases are highlighted with yellow circles, and positions of the two scissile phosphates are denoted with red asterisks (*). (B) Closeup of the covarying residues. A28 (green spheres), G183 (magenta spheres), E29 and E184 (cyan sticks), divalent metal ion cofactors (yellow spheres), scissile phosphates (orange and red sticks) and central 4 bases (gold sticks). (C) Sequence logos of amino acid distribution at the covarying positions from an alignment of 178 single chain meganuclease sequences (23). (D) Overview of nucleotide variation at the I-LtrI and I-OnuI target sites in the mitochondrial rps3 gene of fungi. The central 4 bases of each target site are highlighted with gray rectangle, and shown in detail below.
Each meganuclease has a defined native target site of 22 base pairs, and the enzymes make a double-strand break (DSB) at the central 4 bases, generating 4-nt 3′ overhangs (13). The structural basis of meganuclease-DNA recognition is well understood (5,14,15). Modules of amino acids in anti-parallel β-strands make direct or water-mediated contacts to DNA bases on either side of the central 4 bases, and binding specificity can be modified by directed evolution of these modules.
In contrast, the molecular basis for cleavage specificity at the central 4 bases is unclear. Meganucleases make no direct contacts to the central 4 bases of the target site, and indirect readout of DNA shape and flexibility rather than strict nucleotide identity contributes to recognition and cleavage efficiency (16–18). Profiling of cleavage efficiency on native target sites with central 4 base substitutions has revealed that tolerance to nucleotide variation is different for each meganuclease (16,19,20), likely reflecting evolutionary pressures on meganucleases to compensate for natural variation and genetic drift in their respective target sites. Intolerance to nucleotide variation in the central 4 bases is generally exacerbated in enzymes engineered to bind non-native target sites and thus represents a bottleneck in engineering meganucleases for gene editing or other biotechnological applications (16,17,21,22).
Meganuclease active sites contain conserved aspartate (D) or glutamate (E) residues that coordinate divalent metal ions involved in DNA catalysis, yet the evolutionary and mechanistic implications of this subtle change in residue identity on catalysis or substrate specificity have not been explored. We previously used high-quality structure-guided alignments and computational methods based on mutual information to predict and validate networks of covarying amino acids that impacted meganuclease function (23). Our analyses characterized a covarying network in the meganuclease active site, consisting of the two metal-binding residues (D or E) and two adjacent residues in the LAGLIDADG α-helices (Figure 1). We validated the predicted network by showing that it directly regulates cleavage activity of the meganuclease enzymes I-LtrI and I-OnuI over a ∼100-fold range on their native substrates. The identity of residues in the network correlates strongly with the phylogeny of the meganuclease gene family, suggesting that the active site network coevolves to modulate the fitness landscape of meganuclease activity.
Interestingly, previous screens for up-activity variants of meganucleases engineered against non-native targets identified mutations in residues of the active site network (7,19,24,25), although the coevolutionary significance of these mutations was not fully appreciated at the time. In particular, I-OnuI engineered to cleave a target site in the human monoamine oxidase B gene had poor activity until an E to D substitution (E178D) was introduced in the active site (19).These observations led us to hypothesize that the low activity of meganucleases on substrates with substitutions in the central 4 bases reflects sub-optimal active site-central 4 base combinations that preclude efficient cleavage.
Here, we test this hypothesis using the well-characterized meganucleases I-LtrI and I-OnuI (26). Each of these enzymes has been engineered against different targets for gene editing and gene drive applications, but with varying levels of cleavage efficiency (8,19,27). We screened libraries of 1600 active site variants in the I-LtrI and I-OnuI coevolving network against the native and 67 substrates that differ in the central 4 bases, using a two-plasmid selection system that reports on fitness of meganuclease variants relative to each other on each substrate (23). Overall, we find that the identity of the two metal-binding residues (D or E) has the greatest impact on substrate preference. Biochemical and structural characterization of one predominant variant (E184D) revealed molecular insight into how a single, conservative substitution in the active site impacts indirect DNA readout and substrate preference.
MATERIALS AND METHODS
Oligonucleotides and plasmid libraries
All oligonucleotides were synthesized by Integrated DNA Technologies (IDT), Inc. unless otherwise stated. All plasmid DNA was isolated from Escherichia coli NEB5α cultures grown in Luria Broth (LB) or 2xYT (16 g/l tryptone, 10 g/l yeast extract, 5 g/l NaCl) using an EZ-10 Spin Column Plasmid DNA Kit (Bio Basic Inc.). The I-LtrI and I-OnuI coding regions (codon-optimized for E. coli) were cloned between NcoI and NotI sites of plasmid pEndo. I-LtrI and I-OnuI target sites were cloned between NheI and SacII sites of plasmid pTox for genetic selection assays, or between EcoRI and BamHI sites of plasmid pLitmus28i (Addgene) for in vitro cleavage assays. Individual I-LtrI and I-OnuI active site variants were constructed as previously described (23). I-LtrI and I-OnuI mutagenic libraries were constructed using customized sewing polymerase chain reaction (PCR) strategies. Briefly, open reading frame fragments were amplified from plasmid templates using oligonucleotides containing randomized codon positions (NNS). Reaction products were gel purified and subsequently PCR sewn together using flanking primers containing NcoI and NotI sites, and cloned into pEndo. All libraries were synthesized with 10-fold coverage and a minimum of three independent clones were sequenced (London Regional Genomics Centre) to ensure the correct amino acid positions were mutagenized.
Bacterial two-plasmid functional selection
A previously described E. coli two-plasmid functional selection (23,28,29) was used to screen the activity of all LHE variants and libraries used in this study. For selective growth experiments in liquid culture, 20 ng of LHE variant in pEndo was transformed into 50 μl of chemically competent NovaXGF (Novagen) cells harboring the appropriate pTox plasmid. Transformants were recovered in 2 ml carbohydrate-free 2xYT medium at 37°C in a rotary wheel for 30 min. pEndo expression was induced for 1–4 h using 0.02% arabinose and 100 μg/ml carbenicillin in a rotary wheel at 37°C. Following expression, I-LtrI or I-OnuI cultures were diluted 200-fold into either non-selective (1× M9 salt, 0.8% (w/v) tryptone, 1% (v/v) glycerol, 1 mM MgSO4, 1 mM CaCl2, 0.2% (w/v) thiamine, 100 μg/ml carbenicillin and 0.02% (w/v) L-glucose) or selective media (non-selective media lacking glucose and with the addition of 0.02% (w/v) L-arabinose and 0.1 mM isopropyl beta-D-1-thiogalactopyranoside). Cultures were grown at 37°C for 16 h before cells were pelleted and plasmid DNA was isolated. Codons for the four covarying LHE positions were subsequently amplified from selective and non-selective culture DNA by GoTaq Hotstart PCR (Promega) using customized barcoded primers. Equimolar volumes of each PCR produced were pooled for Illumina sequencing. Solid media selections were performed as above with the following modifications: after the endonuclease expression period (1–4 h), cells were harvested and re-suspended in sterile saline (0.9% wt/vol NaCl), diluted and spread onto non-selective and selective agar plates (selective and non-selective media with 15 g/l agar). Plates were incubated at 37°C for 16–24 h, and the survival percentage was calculated as the ratio of colonies on selective to non-selective plates. A minimum of three biological replicates were performed and background survival was estimated using an empty pEndo vector as a negative control for all selections.
Protein expression and purification
LHE protein variants were cloned between NcoI and NotI sites of pProExHta (Invitrogen and Life Technologies), and the 6x histidine-tagged proteins were over-expressed and purified as previously described (23), with the following modifications. Following the 16 hr Tobacco Etch Virus protease digest at 4°C, I-LtrI variants were purified by cation exchange chromatography using a 5 ml HiTrap SP HP column (GE Healthcare). Peak fractions were pooled and incubated with 50 mM ethylenediaminetetraacetic acid at 4°C for a minimum of 4 h and exchanged into storage buffer (250mM NaCl, Tris–HCl, pH 8.0, 10% [v/v] glycerol and 30 mM CaCl2 ) before concentrating to 5 mg/ml (30ml Amicon filters, Merck) and storage at −80°C.
Crystallization substrates and procedures
I-LtrI ATAC substrate 1: 5′-CAAATGCTCCTATACGACGTTTAGACC-3′, 5′-GGTCTAAACGTCGTATAGGAGCATTTG-3′. I-LtrI ATAC substrate 2: 5′-CAAATGCTCCTATACGACGTTTAGAC-3′, 5′-GGTCTAAACGTCGTATAGGAGCATTT-3′. I-LtrI AACC substrate 1: 5′-CAAATGCTCCTAACCGACGTTTAGACC-3′, 5′-GGTCTAAACGTCGGTTAGGAGCATTTG-3′. I-LtrI AACC substrate 2: 5′-CAAATGCTCCTAACCGACGTTTAGAC-3′, 5′-GGTCTAAACGTCGGTTAGGAGCATTT-3′. I-LtrI ATAC substrate 1 was used for crystallizing structure 6BCE while ATAC substrate 2 was crystallized with structures 6BCF and 6BCG. I-LtrI AACC substrate 1 was crystallized with structures 6BCI and 6BCT while AACC substrate 2 was crystallized with structure 6BCN. Protein preparations were combined with hybridized substrate duplexes in a 1:1.5 ratio (protein:substrate) in the presence of 30 mM CaCl2 and incubated for a minimum of 4h at 4°C to promote pre-cleavage complex formation. Crystallization screens were first performed using the hanging-drop method and commercially available solutions (Wizard II, Ragaku Reagents Inc.). I-LtrI AEGE crystals in complex with the ATAC substrate grew in a 1:1 ratio of protein (5 mg/ml) and precipitant solution (20% [w/v] PEG-2000 MME (Poly(ethylene glycol) methyl ether 2000), 0.1 M Tris–HCl, pH 7.0 and 30 mM CaCl2). I-LtrI variant (AEAE, GEGE and AEGD) crystals in complex with the ATAC substrate grew in a 1:1 ratio of protein (5 mg/ml) and precipitant solution (10% [w/v] PEG-8000, 0.1 M CHES, pH 9.5, 0.2 M NaCl and 30 mM CaCl2). I-LtrI AEGE crystals in complex with the AACC substrate grew in a 1:1 ratio of protein (5 mg/ml) and precipitant solution (2.5 M NaCl, 100 mM NaK phosphate, pH 6.2 and 30 mM CaCl2). I-LtrI AEGD crystals in complex with AACC substrate grew in a 1:1 ratio of protein (5 mg/ml) and precipitant solution (20%[w/v] PEG-2000 MME, 0.1 M Tris–HCl, pH 7.0 and 30 mM CaCl2). All droplets were equilibrated over 1.5 M ammonium sulfate at 18°C, and crystal growth was achieved within 7 days for all constructs.
Structural analysis
Diffraction data were collected using Beam 17-ID at the Advanced Photon Source of Argonne National Labs. Data were collected in quarter degree wedges with an exposure time of 0.0877 s/image. One thousand eighty images representing 270° of rotation or 1440 images representing 360° comprise the dataset (Table 4). Images were indexed and integrated using iMOSFLM. Reflections were scaled and merged using the Aimless and Ctruncate modules from CCP4i (30). Merged reflections were then used for molecular replacement in PHENIX (31). An existing I-LtrI structure (PDB: 3R7P) was used as a search model for all mutants. Corresponding mutations to the protein structure and changes in the central 4 cleavage site were done manually in Coot. Models were refined manually in Coot and using the refine module from Phenix until the Rfree and Rwork factors converged. Structural parameters for DNA substrate structures were analyzed using 3DNA (32).
Table 4.
Summary of crystallographic parameters
| Protein Variant | AEGE | AEAE | GEGE | AEGE | AEGD | AEGD |
|---|---|---|---|---|---|---|
| PDB Accession code | 6BCE | 6BCF | 6BCG | 6BCI | 6BCT | 6BCN |
| Core 4 Sequence | ATAC | ATAC | ATAC | AACC | AACC | ATAC |
| Diffraction source | B-17-ID | B-17-ID | B-17-ID | B-17-ID | B-17-ID | B-17-ID |
| Detector | Dectris 6M | Dectris 6M | Dectris 6M | Dectris 6M | Dectris 6M | Dectris 6M |
| Crystal-detector distance (mm) | 250 | 250 | 250 | 250 | 250 | 250 |
| Rotation range per image (°) | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 | 0.25 |
| Rotation range (°) | 210 | 210 | 210 | 360 | 360 | 360 |
| Exposure time | 0.12 | 0.12 | 0.12 | 0.08 | 0.08 | 0.08 |
| Space Group | C 1 2 1 | P1 | P1 | C 1 2 1 | C 1 2 1 | P1 |
| a,b,c (A) | 115.26, 42.75, 103.38 | 44.01, 66.77, 169.35 | 44.01, 66.77, 169.35 | 117.7, 43.6, 105.6 | 116.2, 43.4, 104.7 | 44.01, 66.77, 169.35 |
| α, β, γ (°) | 90.00, 109.22, 90.00 | 90.02, 89.95, 90.03 | 90.02, 89.95, 90.03 | 90.0, 106.0, 90.0 | 90.0, 107.0, 90.0 | 90.02, 89.95, 90.03 |
| Mosaicity (°) | 0.43 | 0.38 | 0.52 | 0.31 | 0.543 | 0.571 |
| Total no. of reflections | 144 059 | 148 917 | 237 651 | 114 528 | 76 741 | 188 282 |
| Unique reflections | 48 134 | 40 955 | 42 174 | 23 935 | 13 720 | 60 488 |
| Completeness (%) | 99.5 (99.2) | 98.0 (96.9) | 99.9 (98.9) | 100 (99.9) | 100 (100) | 98.0 (97.1) |
| Redundancy | 3.0 (2.8) | 7.2 (2.0) | 5.6 (5.8) | 4.8 (4.5) | 5.6 (5.4) | 3.2 (3.1) |
| I/σ (1) | 8.0 (2.0) | 7.2 (2.0) | 6.5 (2) | 8.9 (2.0) | 7.6 (2.0) | 3.9 (1.3) |
| R merge (%) | 6.2% (44.2%) | 8.0% (48.6%) | 6.5% (33.6%) | 4.7% (44.2%) | 10.6% (59%) | 10.9% (53.4%) |
| Model refinement | ||||||
| Resolution Range | 56.02-2.00 | 66.77-2.92 | 66.58-2.90 | 56.51-2.28 | 56.22-2.73 | 84.49-2.5 |
| R Work | 18.85 | 23.91 | 23.34 | 23.73 | 22.7 | 23.34 |
| R free | 21.45 | 30.86 | 27.36 | 26.83 | 28.16 | 27.36 |
| RMSD bond lengths (Å) | 0.009 | 0.01 | 0.06 | 0.005 | 0.004 | 0.006 |
| RMSD bond angles (Å) | 1.14 | 1.34 | 1.56 | 0.765 | 0.69 | 1.561 |
| Average B factor (Å2) | 36.4 | 74.6 | 45.5 | 64.9 | 86.1 | 45.5 |
| Ramachandran Outliers (%) | 0 | 0 | 0.1 | 0 | 0 | 0.4 |
| Rotamer Outliers (%) | 1.6 | 1.8 | 1.5 | 3.4 | 0.4 | 2.7 |
| ClashScore | 3.83 | 10.42 | 12.78 | 4.55 | 5.69 | 10.8 |
Endonuclease assays
Endonuclease assays were performed as previously reported (23). Briefly, reactions were performed using six different protein concentrations and 5 nM plasmid substrate at 37°C. Plasmid species were separated using 0.9% (w/v) agarose-TBE gel electrophoresis and product formed was calculated as the intensity of the linear product band divided by the sum of the three reactants (supercoiled substrate, nicked plasmid and linear product). Initial rates from each of the six time-course experiments were then plotted against enzyme concentration and fit to a Michaelis–Menten model to determine the parameters kcat* and Km* (33). All experiments were repeated in triplicate.
Product enrichment assays
I-LtrI and I-OnuI targets with randomized C4 regions (I-LtrI 5′-AATGCTCCTNNNNGACGTTTAG-3′; I-OnuI 5′-TTTCCACTTNNNNAACCTTTTA-3′) were cloned between EcoRI and BamHI sites of pLitmus28i (Addgene) and used as inputs for time-course endonuclease assays [500 nM protein, 5 nM DNA substrate, 100 mM NaCl, 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol (DTT) at 37°] with purified I-LtrI or I-OnuI variants. Reaction products were separated from unreacted substrates using 0.9% (w/v) agarose-TBE gel electrophoresis at 3.5 V/cm for 75 min, and linear product bands excised from the gel matrix and purified using EZ-10 Spin Column DNA Gel Extraction Kits (Bio Basic Inc.). Linearized plasmid DNA was re-circularized using T4 DNA Ligase (New England Biolabs), and the re-circularized products and input libraries were amplified using GoTaq Hotstart PCR (Promega) with customized barcoding primers flanking the recognition site. Equimolar volumes of each sample were pooled and subject to high-throughput sequencing (n = 5).
Sequencing and data analysis
High-throughput paired-end sequencing used an Illumina NextSeq at the London Regional Genomics Centre (London, ON), or an Illumina HiSeq at The Centre for Applied Genomics (The Hospital for Sick Children, Toronto, ON). Unique 6- or 8-bp barcode sequences were used to group reads into the appropriate sample (34). Read counts per feature were identified using a customized Perl script. Specifically, functional selection-based reads were parsed for the presence of Asp (D) or Glu (E) at positions 29 and 184, while reads from the in vitro central 4 substrate preference assays were parsed for the correct I-LtrI or I-OnuI target sequence between nucleotide positions −11 and −4. The relative abundance of co-evolving network variants (in the selected versus non-selected condition) and enriched central 4 products (in linearized pool versus input library) was determined using the ALDEx2 R package (ANOVA-Like Differential Expression 2.0) (35,36). Each Monte-Carlo instance was converted to a log-ratio using the IQLR method in ALDEx2 (35); this is referred to as the *LR transformation. This approach, known as compositional data analysis (37), results in *LR and related log-ratio transformations where the data are on a linear-scale and now can be used with standard statistical approaches (37–39), almost without restriction. All hypothesis tests were corrected for false discovery rate (FDR) using the Benjamini and Hochberg (40) adjusted Welch’s t-tests, and ALDEx2 reports the expected value of these tests (35). An expected q-value of 0.05 was used to assess statistical significance. This approach has essentially no false positive identifications while losing little, if any sensitivity (41–44). We also used a standardized effect size (42,45,46) and distributional overlap outputs from ALDEx2 to assess biological significance and reproducibility of the data. The standardized effect is the expected value of the difference between groups divided by the dispersion within groups (35,42). Features were identified as significantly, and biologically distinct, if they had an absolute magnitude of change >log2(2), an absolute effect size >2 (meaning the within group dispersion was less than half the between group difference), an overlap of >1% (meaning that the confusion between sample groups was >1%), and a q-value >0.05. Similar cutoffs using the same tool have proven to give robust conclusions even with very small sample sizes (47). Plots were generated with the R package ggplot2 (48).
RESULTS
Identification of meagnuclaese variants active on substrates with central 4base substitutions
We used a well-described two-plasmid functional assay where survival of E. coli depends on meganuclease expression from pEndo cleaving a target site cloned into a toxic plasmid (pTox) expressing the DNA gyrase toxin ccdB (Figure 2A) (23,29). Non-permissive meganuclease–substrate combinations do not cleave the pTox, and are bacteriostatic due to expression of ccdB. This assay can be performed with solid or liquid media, and was previously used to assess fitness effects of mutations in the co-evolving catalytic network of three different meganucleases on their cognate substrates (23). When tested with substrates that differed from the cognate substrates in the central 4 bases, I-LtrI or I-OnuI survival ranged from ∼0.01 to ∼50%, whereas the cognate substrates promoted ∼100% survival (Figure 2B). Thus, any meganuclease variant with activity on a central 4 substrate poorly cleaved by the wild-type enzyme would exhibit a significant relative fitness advantage.
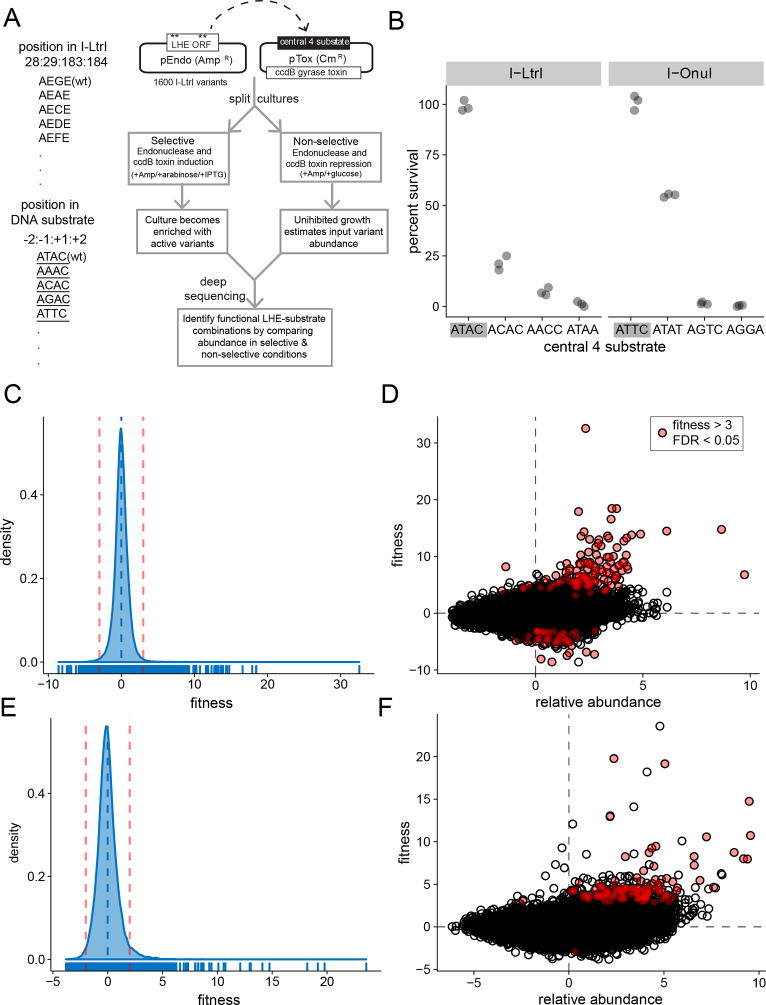
Figure 2.
Experimental workflow and analyses. (A) Schematic of two-plasmid functional selection and nomenclature for naming protein variants and DNA substrates. Note that DNA substrates are distinguished from protein variants by an underline (i.e. ATAC). (B) Results of two-plasmid assay on solid media with wild-type I-LtrI and I-OnuI against cognate ATAC and ATTC substrates (gray rectangles), and substrates differing in the central 4 bases. Data points are derived from three independent biological replicates. (C) Histogram of fitness derived from deep sequencing data for all I-LtrI variants on all tested central 4 substrates (78 400 data points). The vertical dashed blue line represents the mean fitness, and the vertical dashed red lines represent the 99% confidence intervals for the data. The x-axis is log2 scale. (D) Plot of variant relative abundance versus variant fitness for all I-LtrI data. Red dots are variants with fitness values >3 and a FDR of <0.05. Axes are log2 scale. (E) and (F) Fitness histogram and variant abundance versus fitness plots for the I-OnuI experiments. Plots are labeled as in panels (C) and (D).
We screened libraries of 1600 active site variants in I-LtrI and I-OnuI that were generated by randomizing residue positions 28 and 183 to all 20 amino acids, while constraining the metal-binding residues at positions 29 and 184 to D or E (I-LtrI numbering will be used for simplicity) (23). DNA substrates are identified by the central 4 bases and are underlined to distinguish them from protein variants (Figure 2A). Using this nomenclature, the I-LtrI active site would be AEGE and the native DNA substrate ATTC. This screen assessed the fitness landscape of the meganuclease active site on 67 substrates with substitutions in the central 4 bases, and the 2 cognate substrates (ATAC and ATTC) (Table 1). Following outgrowth of the replicate experimental (selective) or control (non-selective) samples, deep sequencing was used to determine relative meganuclease variant abundance for each substrate tested. This approach allowed us to generate a fitness value for each variant that is the dimensionless effect size representing the magnitude of change between experiment and control conditions scaled by the variance within each condition (42,45,46). The effect statistic directly addresses the biologically relevant question of what is reproducibly different between the experimental and control conditions. We considered variants as significant if fitness values of individual variants were greater than the 99% confidence interval of the aggregate I-LtrI or I-OnuI data (Figure 2C and E). Because we performed multiple replicates of experimental and control conditions for each library–substrate combination, a FDR of >0.05 was also applied to identify significant variants (red-filled circles in Figure 2D and F). Variants with the highest fitness values on each substrate are presented in Table 1. The major findings from these experiments are:
I-LtrI variants with high reproducible fitness were identified on 6/49 (12%) of the substrates tested (AAAC, ATAC, ATCC, CTAC, GATT, TTTC), and I-OnuI variants on 4/19 (26%) of the substrates tested (AATC, AGTC, ATAT and TTTC).
The wild-type active site network (AEGE) was identified as the best variant for only a single substrate (TTTC for I-OnuI).
The I-LtrI AEGD variant was identified as the best variant on two substrates (AAAC and ATCC), and as the best I-OnuI variant on three substrates (AATC, AGTC and ATAT).
In total, 55 unique variants with active site residue combinations not found in known meganucleases were identified. Some of these variants had high fitness values, including I-LtrI TDGD on the TTTC substrate and I-LtrI QDWD on the GATT substrate.
Table 1.
Summary of I-LtrI and I-OnuI selection experiments
| Protein | Substrate | nt. diffs | AEGE fitness | Best variant | Best variant fitness | Found in alignment? |
|---|---|---|---|---|---|---|
| I-LtrI | ATAC (wt) | 0 | 13.93 | GEAD | 32.56 * | No |
| I-LtrI | AAAC | 1 | 3.7 | AEGD | 6.78 * | Yes |
| I-LtrI | ACAC | 1 | 0.65 | LEVD | 1.96 | No |
| I-LtrI | AGAC | 1 | −0.89 | ADRE | 2.34 | No |
| I-LtrI | ATAA | 1 | 2.03 | SDGD | 2.23 | Yes |
| I-LtrI | ATAT | 1 | 1.96 | RDWE | 4.85 | No |
| I-LtrI | ATCC | 1 | 4.69 | AEGD | 13.78 * | Yes |
| I-LtrI | CTAC | 1 | 1.89 | QDGD | 7.17 * | No |
| I-LtrI | AACC | 2 | 1.69 | AETD | 2.17 | No |
| I-LtrI | AAGC | 2 | 1.3 | NEIE | 2.3 | No |
| I-LtrI | AATC | 2 | 1.37 | AELD | 1.82 | No |
| I-LtrI | ACAA | 2 | 1.4 | PEGE | 4.82 | No |
| I-LtrI | ACCC | 2 | 1.25 | QEND | 4.97 | No |
| I-LtrI | ACTC | 2 | 0.05 | WEPE | 5.38 | No |
| I-LtrI | AGAA | 2 | 0.35 | VEND | 2.16 | No |
| I-LtrI | AGTC | 2 | 1.42 | TDGD | 3.31 | No |
| I-LtrI | ATCA | 2 | −0.83 | REYE | 3.79 | No |
| I-LtrI | ATGT | 2 | −0.23 | PDGD | 1.91 | No |
| I-LtrI | ATTT | 2 | −0.02 | QDAD | 1.5 | No |
| I-LtrI | GTCC | 2 | 1.78 | VDKE | 3.91 | No |
| I-LtrI | GTGC | 2 | 1.12 | QDDE | 2.73 | No |
| I-LtrI | TTAA | 2 | 0.5 | VEHD | 2.78 | No |
| I-LtrI | TTGC | 2 | 0.77 | REPD | 2.3 | No |
| I-LtrI | TTTC | 2 | 10.23 | TDGD | 18.46 * | No |
| I-LtrI | AACA | 3 | −0.2 | WETD | 3.46 | No |
| I-LtrI | AATT | 3 | −0.06 | KDTE | 2.16 | No |
| I-LtrI | ACGT | 3 | −0.07 | VDAD | 3.49 | No |
| I-LtrI | AGCG | 3 | −0.85 | RDEE | 1.33 | No |
| I-LtrI | AGGA | 3 | 1.17 | ADAE | 3.74 | No |
| I-LtrI | AGGG | 3 | −0.79 | PECE | 2.75 | No |
| I-LtrI | AGGT | 3 | −1.24 | ADRE | 9.21 | No |
| I-LtrI | CAAT | 3 | −0.2 | DEAD | 4.56 | No |
| I-LtrI | CAGC | 3 | 0.45 | SESE | 5.06 | No |
| I-LtrI | CGCC | 3 | 1.51 | DDGE | 3.49 | No |
| I-LtrI | CGGC | 3 | 0 | IEVE | 5.42 | No |
| I-LtrI | GAAT | 3 | 0.58 | GEAE | 3.04 | Yes |
| I-LtrI | TCCC | 3 | 2.67 | KEGE | 4.93 | No |
| I-LtrI | TTTT | 3 | 1.67 | HDRD | 6.39 | No |
| I-LtrI | CAGG | 4 | 1.41 | IDAE | 3.94 | No |
| I-LtrI | CATA | 4 | 0.01 | TERD | 4.97 | No |
| I-LtrI | CGCG | 4 | −0.88 | PERD | 3.07 | No |
| I-LtrI | CGCT | 4 | 1.46 | IDGE | 4.03 | No |
| I-LtrI | CGGA | 4 | −2.79 | EDSE | 3.48 | No |
| I-LtrI | CGGG | 4 | −0.71 | PEDE | 5.08 | No |
| I-LtrI | CGTG | 4 | −0.2 | KESE | 5.73 | No |
| I-LtrI | GATT | 4 | 0.78 | QDWD | 8.18 * | No |
| I-LtrI | GGCT | 4 | −0.23 | SDPE | 2.64 | No |
| I-LtrI | TCCG | 4 | −1.38 | EEAE | 8.36 | No |
| I-LtrI | TGCG | 4 | 1.11 | MDAE | 5.27 | No |
| I-OnuI | ATTC (wt) | 0 | 10.09 | SEGE | 23.58 | No |
| I-OnuI | AATC | 1 | 5.46 | AEGD | 14.75 * | Yes |
| I-OnuI | AGTC | 1 | 3.41 | AEGD | 8.74 * | Yes |
| I-OnuI | ATGC | 1 | 2.65 | AEIE | 3.11 | No |
| I-OnuI | ATTA | 1 | 2.27 | AENE | 3.74 | No |
| I-OnuI | ATTG | 1 | 2.67 | AERE | 4.61 | No |
| I-OnuI | ATTT | 1 | 8.26 | AEGD | 10.57 | Yes |
| I-OnuI | TTTC | 1 | 8 | AEGE | 8 * | Yes |
| I-OnuI | AAAC | 2 | 1.35 | SEGD | 3.6 | No |
| I-OnuI | AAGC | 2 | 1.63 | IDGD | 4 | No |
| I-OnuI | ATAG | 2 | 1.26 | AEWD | 3.94 | No |
| I-OnuI | ATAT | 2 | 4.65 | AEGD | 10.73 * | Yes |
| I-OnuI | ATCA | 2 | 1.05 | IECD | 2.76 | No |
| I-OnuI | ATCG | 2 | 3.21 | AELD | 5.94 | No |
| I-OnuI | CCTC | 2 | 3.15 | AERD | 3.81 | No |
| I-OnuI | GGTC | 2 | 2.14 | AECD | 3.11 | No |
| I-OnuI | TTGC | 2 | 2.81 | AEDD | 3.83 | No |
| I-OnuI | AGGA | 3 | 6.26 | AEGE | 6.26 | Yes |
| I-OnuI | AGGG | 3 | 0.42 | QDED | 2.3 | No |
Asterisks (*) indicate variants with FDR values <0.05 and true positive confidence interval is >95%. Found in alignment indicates whether the amino acid sequence of the best variants was found in the alignment of meganuclease sequences used to generate the sequence logos plot in Figure 1C.
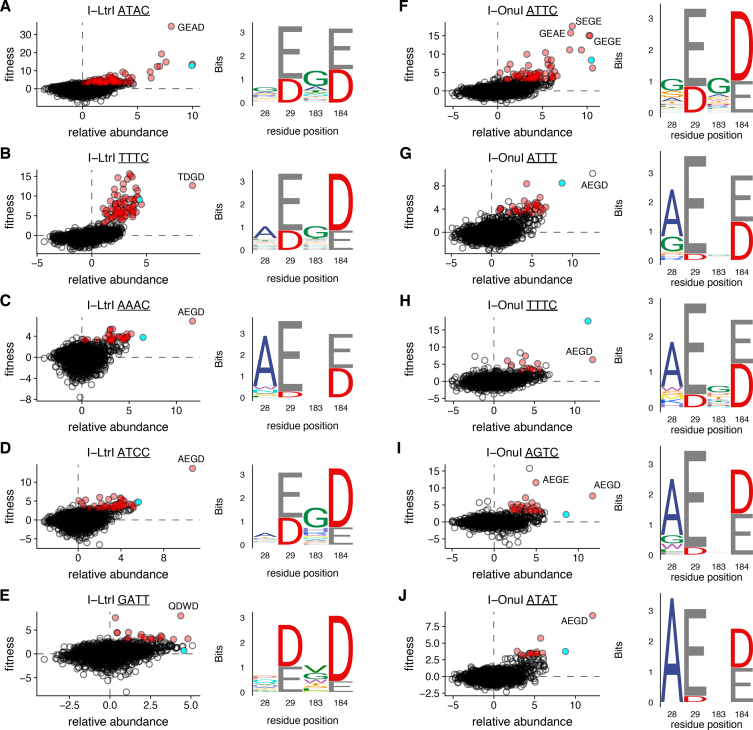
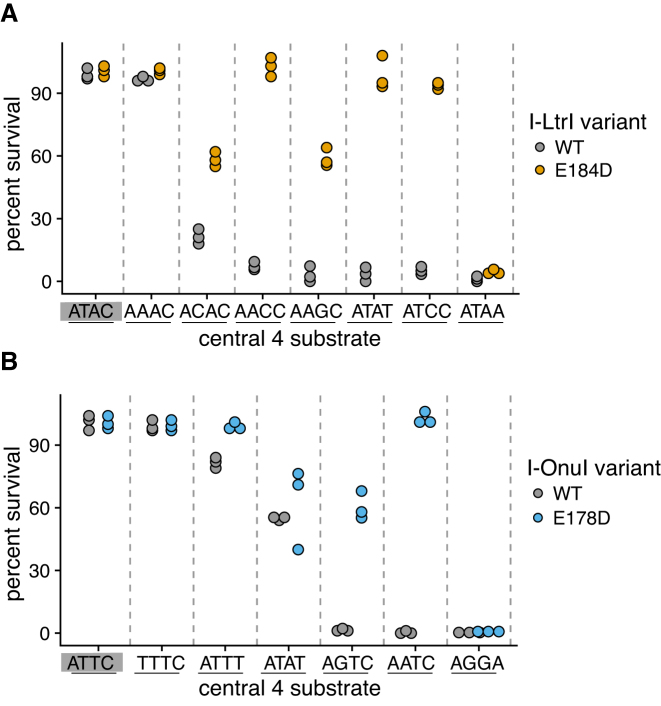
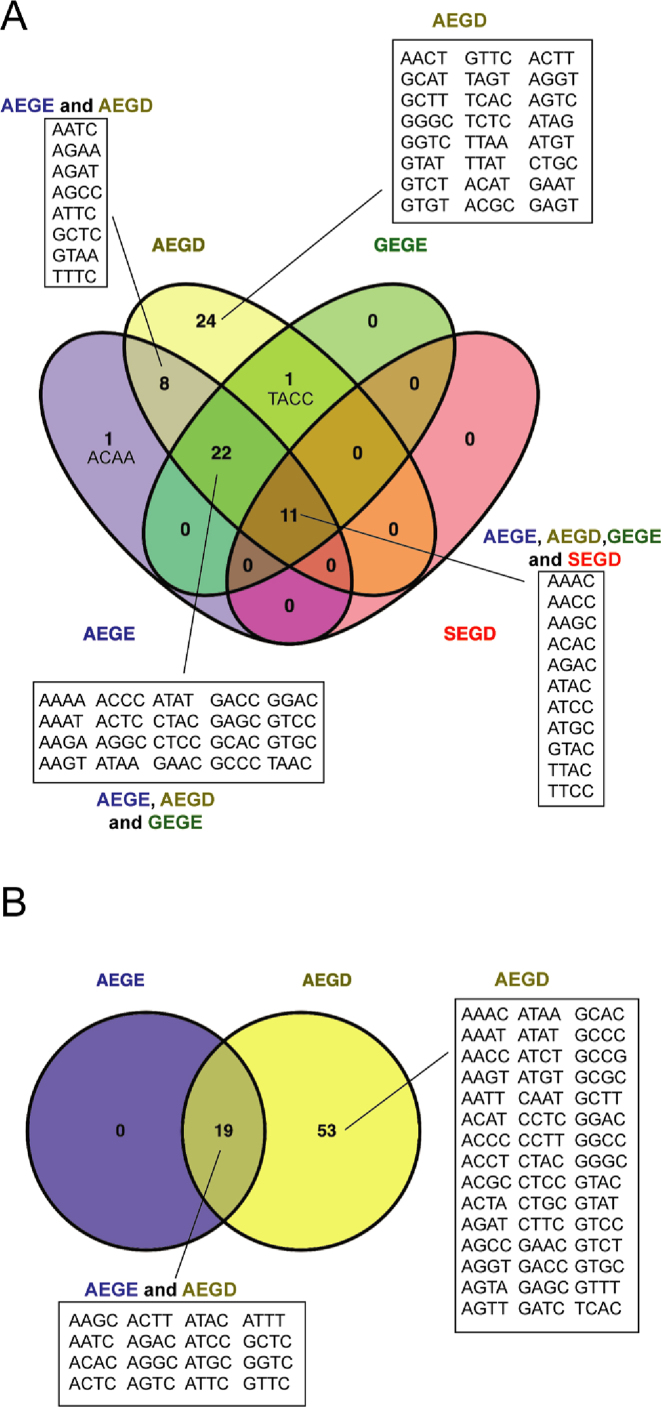
One unexpected finding from our study was the diversity of changes in the I-LtrI and I-OnuI active sites that supported activity on a variety of substrates. These active site variants were distinct from those isolated on the cognate substrates (Figure 3B–J and Supplementary Figures S1 and S2). Notable differences in active site residue identity included changes in preference for metal-ion binding residues (E or D) at positions 29 and 184 and a greater diversity of permissible residues at position 183. We also identified that I-LtrI and I-OnuI variants with residue combinations not found in known meganucleases exhibited robust fitness on a number of substrates (Table 1). Interestingly, the AEGD variant was repeatedly identified as the variant with the highest fitness on five substrates for I-LtrI and I-OnuI (Table 1). We confirmed this observation by showing that the I-LtrI and I-OnuI AEGD variants had higher survival than the wild-type proteins when individually tested against a number of substrates using the two-plasmid functional assay (Figure 4).
Figure 3.
Examples of co-evolving residue variability in I-LtrI or I-OnuI variants selected on different DNA substrates. For each indicated DNA substrate, plots are relative abundance versus variant fitness, with red dots representing significant variants (fitness >3 and FDR of <0.05) and green dots representing the wild-type enzymes. Please note that the x- and y-axis scale differ with each experiment. Sequences logos to the right are generated from significant variants. Panels (A–E) are from I-LtrI experiments, and panels (F–J) are from I-OnuI experiments. Residue positions are labeled according to I-LtrI numbering. Individual variants are identified by amino acid identity.
Figure 4.
Verification of activity on different central 4 substrates for the (A) I-LtrI and (B) I-OnuI AEGD and wild-type proteins. Data points are three independent biological replicates. The cognate I-LtrI ATAC and I-OnuI ATTC substrates are highlighted by a gray rectangle.
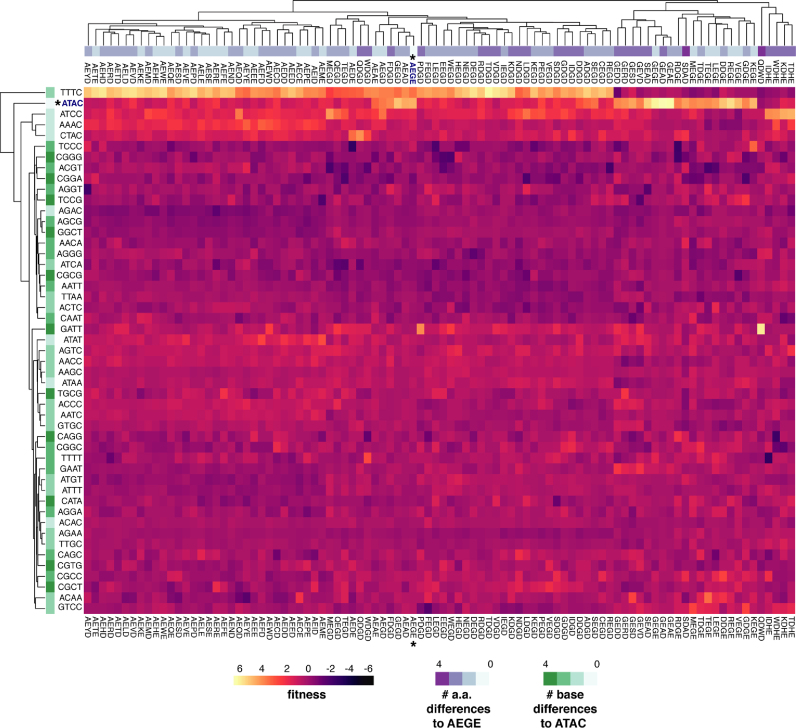
To visualize the effect of substitutions in the co-evolving network on fitness, we plotted the fitness landscape of the most active I-LtrI and I-OnuI variants (fitness >3 and FDR <0.05) as a heatmap for all 49 I-LtrI (Figure 5 and Supplementary Figure S3) and 19 I-OnuI substrates (Supplementary Figures S4 and S5). This global overview reinforced the findings that only a small number of substrates with few nucleotide differences to the cognate ATAC and ATTC substrates supported activity. Of particular interest were I-LtrI variants with robust fitness on the TTTC substrate but poor fitness on the ATAC substrates, and vice-versa (for example, GEGE, GEAD and GEAE). A number of variants had very narrow preference, including the I-LtrI QDWD variant for the GATT substrate (fitness 8.18, Table 1 and Figure 5), but with fitness values ranging from −1.5 to 1.2 for all other substrates. Similar trends were seen with I-OnuI (Supplementary Figure S5), including a set of variants (GEGD, GEAD, SEGD, GEAE, GEGE and SDGE) that were specific for the cognate ATTC substrate, and showing poor activity on all other substrates.
Figure 5.
Heatmap of I-LtrI variant fitness. Variants with fitness >3 and FDR of <0.05 were identified, and fitness values for that variant on all remaining substrates were collated and plotted as a heatmap. The number of base changes for each substrate relative to the cognate ATAC or ATTC substrates are indicated in the vertical (row) heatmap, while number of amino acid changes for each protein variant relative to the wild-type AEGE protein are indicated in the horizontal (column) heatmap.
In summary, the in vivo experiments revealed that many I-LtrI and I-OnuI variants are active against the cognate substrates and substrates that differed by 1 or 2 bases. We noted that E:E and E:D pairs of metal-binding residues at positions 29 and 184 were more active in the I-LtrI and I-OnuI scaffolds than D:E or D:D metal-binding residue pairs. For example, the E184D substitution (typically in the AEGD variant) was enriched against multiple central 4 substrates. In addition, a number of I-LtrI and I-OnuI variants were isolated with low but reproducible fitness values on non-cognate substrates on which the wild-type enzymes showed no activity. Interestingly, many of these residue combinations are not present in known meganuclease family members, possibly indicating ascertainment bias of natural meganuclease sequences, or that these residue combinations represent unstable evolutionary intermediates that only serve as starting scaffolds for further optimization.
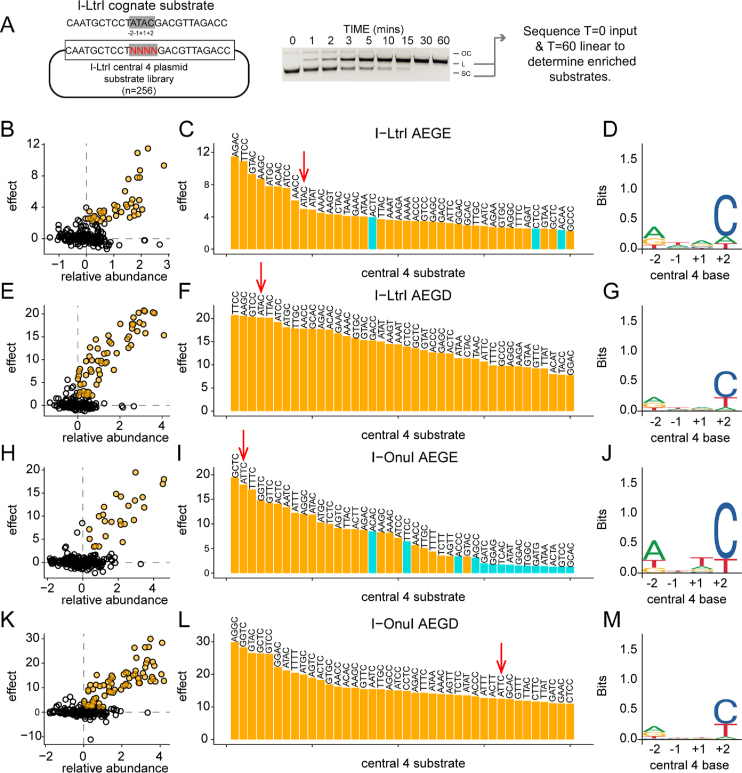
Simultaneous assays on all 256 central 4 substrates reveals broadened cleavage preference of the E184D substitution
We devised a high-throughput in vitro competition assay to profile purified meganuclase variants on all central 4 substrates simultaneously (Figure 6). This assay was used to provide a more global overview of whether the I-LtrI and I-OnuI network variants impacted cleavage preference. I-LtrI and I-OnuI target sites with randomized central 4 regions were cloned to generate plasmid substrate libraries, each with 256 possible combinations of central 4 bases (Figure 6A). These libraries were incubated with purified proteins for time-course cleavage assays, and supercoiled substrate was separated from nicked and linearized reaction products by gel electrophoresis. This experimental strategy enabled the identification of cleavable central 4 substrates by isolating and deep sequencing both the linear products and input libraries to determine enrichment values per substrate (Dataset S3 and S4, Supplementary Table S4 and S5).
Figure 6.
Profiling specificity using in vitro enrichment on 256 central 4 substrates.Substrate specificity of I-LtrI and I-OnuI proteins. (A) Experimental setup, with schematic of plasmid substrate containing the randomized I-LtrI central 4 target sequence (left), an example of a cleavage assay (middle), and downstream analysis (right). Panels (B–D) are enrichment results from I-LtrI AEGE (wt) protein with plot of relative abundance versus substrate effect size with significant substrates (effect >2 and FDR <0.05) colored yellow (on left), a ranked barplot of significant substrates, and a logos plot of significantly enriched substrates (yellow bars from barplot). Panels (E–G), I-LtrI AEGD. Panels (H–J), I-OnuI AEGE (wt). Panels (K–M), I-OnuI AEGD. Red arrows indicate the cognate substrate (ATAC for I-LtrI and ATTC for I-OnuI).
We first examined the cleavage profile of the I-LtrI AEGE (wt) and AEGD proteins, using the effect statistic to determine significantly enriched substrates (Figure 6B and E) that were plotted in rank order to visualize differences (Figure 6C and F). Interestingly, the cognate ATAC substrate was not the preferred substrate. Sequence logo (49) plots constructed from significantly enriched substrates indicated that information content readout by the AEGD variant at positions +1 and +2 of the central 4 bases was reduced relative to the wild-type AEGE enzyme (Figure 6D and G), consistent with a broader cleavage profile indicated from the in vivo experiments. We also examined the cleavage profile of two other network variants, GEGE (A28G) and SEGD (A28S/E184D) that were enriched against some central 4 substrates in vivo (Figure 5). We previously showed that the GEGE variant is more catalytically efficient against the cognate ATAC substrate than wild-type I-LtrI, and that SEGD has catalytic activity comparable to wild-type I-LtrI on the cognate ATAC sequence (23). Sequence logo plots showed that both the GEGE and SEGD variants had narrower cleavage profiles than the AEGE and AEGD variants as evidenced by an increased information content readout, particularly at position +2 of the central 4 bases (Supplementary Figure S6). Comparison of substrate preference for the 4 I-LtrI variants revealed 11 substrates were significantly enriched by all variants, whereas the AEGD variant significantly enriched 24 unique substrates relative to other enzymes (Figure 7A). The enrichment profiles of the I-OnuI wild-type AEGE and AEGD enzymes revealed a similar pattern in that information readout at the +1 and +2 positions of the central 4 bases was lower for the AEGD variant (Figure 6J and M). This reduction in information readout resulted in a broadening of cleavage preference, with the AEGD variant cleaving 53 central 4 substrates not significantly cleaved by the wild-type enzyme (Figure 7B). Our cleavage preference profiling of wild-type I-LtrI and I-OnuI differs from previous nucleotide-by-nucleotide profiling of central 4 base preferences (19), notably in the middle two positions (+1/−1) where our data indicate little information readout by both enzymes. These differences may indicate that indirect readout by I-LtrI and I-OnuI of the central 4 bases (and thus central 4 preference) is more sensitive to multiple nucleotide substitutions assayed in our study (which would have a greater effect on DNA structure), as opposed to single substitutions that have compartively smaller effects on indirect readout and cleavage efficiency.
Figure 7.
Substrate specificity of I-LtrI and I-OnuI proteins. (A) Substrate preference of I-LtrI proteins shown as a Venn diagram. (B) Substrate preference of I-OnuI proteins shown as a Venn diagram.
Increases in kcat* allows E184D to overcome defects in binding
To provide further insight into how the E184D substitution broadens the cleavage profile of I-LtrI, we determined single-turnover kinetic constants for the AEGE and AEGD I-LtrI variants on the cognate ATAC and AACC substrates (Table 2). The AACC substrate was chosen because both enzymes were active on this substrate, allowing us to discern mechanistic information from differences in kinetic constants (as opposed to little kinetic information that would be obtained from poorly cleaved substrates). For the AEGE wild-type enzyme, Vmax and kcat* were similar on both substrates, but a ∼10-fold increase in KM* resulted in a ∼10-fold reduction in catalytic efficiency (kcat*/KM*) against the AACC substrate. Interestingly, while KM* for the AEGD variant was similar to that of the wild-type AEGE protein on the AACC substrate, a higher catalytic efficiency of the AEGD variant was attributed to an ∼3-fold higher kcat* that compensated for reduction in binding. This trend was also observed for the AEGD variant on the ATAC native substrate. The higher kcat* values for the AEGD variant help explain the in vivo experiments where faster cleavage of the pTox-AACC substrate plasmid would result in higher enrichment values relative to the wild-type AEGE enzyme.
Table 2.
Single turnover kinetics for I-LtrI AEGE and AEGD variants on the ATAC and AACC substrates.
| Protein | Substrate | Vmax (nM/s) | k cat (nmol/sec) | K m (nM) | Efficiency (kcat/Km) |
|---|---|---|---|---|---|
| AEGE | ATAC | 0.29 | 0.059 ± 0.0021 | 48 ± 10 | 0.0012 |
| AEGE | AACC | 0.29 | 0.058 ± 0.0077 | 458 ± 138 | 0.00013 |
| AEGD | ATAC | 1.06 | 0.21 ± 0.009 | 163 ± 26 | 0.0013 |
| AEGD | AACC | 0.77 | 0.16 ± 0.027 | 561 ± 178 | 0.00028 |
Meganucleases generate a DSB by two sequential nicking reactions that hydrolyze the phosphodiester backbone of each DNA strand (25,50). The individual first-order rate constants can be determined in vitro from the appearance of nicked intermediate (k1) and linear product (k2) when using supercoiled substrates that contain the I-LtrI target site with either the cognate ATAC or AACC central 4 sequences (Table 3). For the AEGE wild-type I-LtrI protein, the k2 rate was affected most by the AACC substrate relative to the cognate ATAC substrate. In contrast, the k2 rate constant for the I-LtrI AEGD variant was similar for both the ATAC and AACC substrates, indicating that the E184D substitution enhances second-strand nicking of non-cognate substrates relative to the wild-type AEGE protein.
Table 3.
Rate constants for first (k1) and second (k2) strand nicking reactions.
| Protein | Substrate | k 1 (s−1) | k 2 (s−1) |
|---|---|---|---|
| AEGE | ATAC | 0.0040 ± 0.0007 | 0.021 ± 0.0051 |
| AEGE | AACC | 0.0057 ± 0.0013 | 0.017 ± 0.0023 |
| AEGD | ATAC | 0.016 ± 0.0009 | 0.028 ± 0.0074 |
| AEGD | AACC | 0.010 ± 0.0031 | 0.083 ± 0.016 |
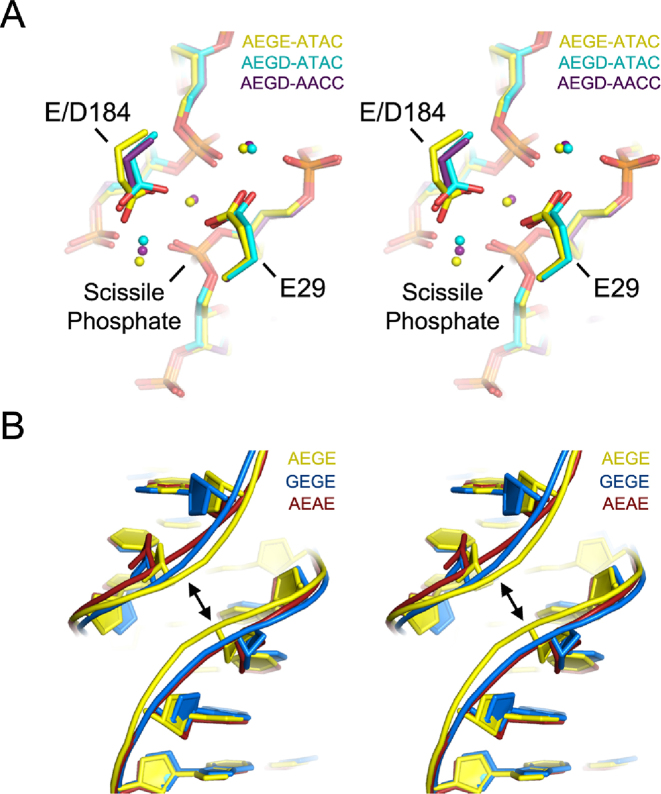
Co-evolving network mutations affect substrate structure and divalent metal coordination within the I-LtrI active site
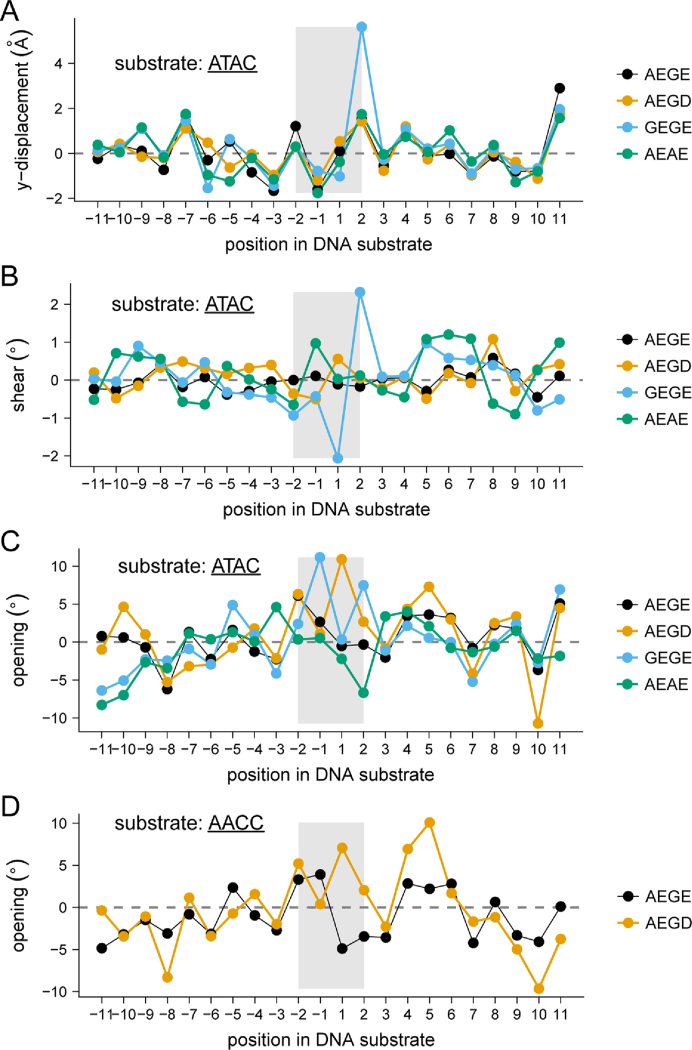
To provide structural insight into differences in catalytic efficiency between the wild-type and I-LtrI variants, we solved pre-cleavage crystal structures of the I-LtrI AEGE (wild-type), GEGE (A28G), AEAE (G183A) and AEGD (E184D) proteins in complex with the cognate ATAC substrate, and the AEGE and AEGD proteins in complex with the AACC substrate (Table 4). Substituting the metal-binding residue at position 184 from E to D resulted in changes in side chain positioning for both the ATAC and ATAC substrates without altering DNA structure (Figure 8A). Repositioning of metal ions was most pronounced in the metal ion immediately adjacent to the scissile phosphate (Figure 8A), suggesting the positioning of catalytic metal ions is an essential factor for governing substrate cleavage specificity. Interestingly, substitutions in the non-metal binding residues of the co-evolving network were found to have less effect on metal ion positioning (for the GEGE and AEAE structures), but substantially changed DNA structure (Figure 8B). In particular, the minor groove widths of the centre of the ATAC substrates were widened to a greater extent in GEGE (5.7 Å) and AEAE (6.9 Å) compared to AEGE (5 Å) and AEGD (5 Å) (Figure 9). Analysis of the AACC substrate in the AEGE and AEGD complexes showed no changes in minor groove width (Figure 9). A detailed summary of DNA structural changes is listed in Supplementary Figures S7 and S8. The central 4 bases in the ATAC substrate adopted a closed conformation (−6.68°) when bound by the poorly active AEAE variant (Figure 9). Bases within the AACC substrate adjacent to the first scissile phosphate were open in both AEGE (3.31°) and AEGD (5.21°), while only AEGD (2.05°) conferred an open structure to bases around the second scissile phosphate (Figure 9). These increases in central 4 base pair opening of the AEGD protein correlated with higher activity of the AEGD variant on the ATAC and AACC substrates relative to the wild-type AEGE protein.
Figure 8.
Structural analyses of I-LtrI variants on the cognate ATAC and AACC substrates. (A). Stereo representation of positions 29 and 184 and DNA substrate of the AEGE-ATAC (yellow), AEGD-ATAC (cyan) and AEGD-AEGD protein–DNA complexes. Variants crystallized with the ATAC and AACC substrates. Dots represent metal ions and are color-coded according to the protein–DNA complex. (B). Stereo representation of showing changes in minor groove width in protein–DNA complexes with I-LtrI variants with substitutions in the non-metal binding residues positions (28 and 183).
Figure 9.
Nucleic acid parameters for protein–DNA complexes. Shown for wild-type I-LtrI AEGE, I-LtrI AEGD, I-LtrI GEGE and I-LtrI AEAE in complex with the cognate ATAC substrate are base pair shear angle (panel (A)), y-displacement ((B)), opening angle (panel (C)). Base pair opening angle (panel (D)) is also displayed for wild-type I-LtrI and I-LtrI E184D against the non-cognate AACC substrate. The nucleotide position is on the x-axis, and the central 4 bases are highlighted by a gray rectangle.
DISCUSSION
In this study, we explored the relationship between amino acid identity in the meganuclease active site and cleavage preference in the central 4 bases of the target site. In general, we find that the wild-type active site residues are not optimized for cleavage of many substrates in one-time cleavage experiments. Notably, residue combinations that are rare or not observed in known meganucleases support activity on substrates poorly cleaved by I-LtrI and I-OnuI. The structural consequences of these substitutions include changes in the indirect readout of the central 4 bases, with concomitant changes in cleavage preference that are specific for each active site variant. Collectively, our data suggest that the residue distribution in known meganuclease active sites resulted from evolutionary pressures that balanced specificity versus activity rather than selection for the optimal chemical solution.
How does the E184D substitution enhance cleavage?
Indirect readout of DNA shape and flexibility rather than strict nucleotide identity plays a critical role in DNA recognition and cleavage efficiency for a number of DNA-binding proteins and site-specific DNA endonucleases (16,18,51–57). Indirect readout of the central 4 bases largely determines meganuclease cleavage efficiency and is influenced by energetics of DNA twisting and bending (16–18). Notably, GC-rich central 4 sequences are poorly cleaved by characterized meganucleases, possibly because GC base steps increase stacking energy and decrease DNA flexibility. Structural studies of the meganuclease I-CreI with substrates containing non-cleavable central 4 sequences with central GC bases (in positions −1/+1) has revealed widening of the DNA minor groove near the scissile phosphates (18), whereas Y-displacement of bases that caused DNA backbone changes and loss of coordinated divalent metal ion was observed in co-crystals of I-SmaI and non-cleavable substrates (16). Our data generally agree with these results, showing that substrates enriched by the wild-type I-OnuI and I-LtrI proteins are mostly AT-rich. In contrast, the AEGD variant (with the E184D substitution) enriched more substrates with C at position +2 of the central 4 bases (Figure 7). This C+2 preference could in part be explained by a direct contact made by R311 of I-LtrI to the bottom strand G at the +2 position of the wild-type complex (Supplementary Figure S9). This contact could stabilize the pre-cleavage complex that restricts conformational flexibility of DNA necessary for limiting substrate cleavage specificity. The R311-C+2 contact was not observed in the non-wild type structures determined here, nor the previously reported post-cleavage complex (19). Interestingly, the greatest increases in substrate enrichment for the I-LtrI AEGD variant were observed on central 4 base substrates with higher predicted flexibility, suggesting that flexibility is a rate limiting step for LHE cleavage and that the E184D substitution contributes to de-stabilization of the protein-DNA structure that normally restricts cleavage specificity.
Previous studies with restriction enzymes have shown that substitutions in DNA or protein residues involved in indirect readout had larger impacts on DNA cleavage than on DNA binding (52). This does not appear to be the case with the I-LtrI E184D mutant where Km is affected more than kcat (Table 2). This finding suggests that negative impacts on DNA binding are compensated for by corrections in active site alignment with substrates that overcome energetic barriers in DNA structure that prevent efficient cleavage by the wild-type protein. Increases in base pair opening at positions +1/+2 that are adjacent to the top-strand scissile phosphate with the I-LtrI AEGD (E184D) variant and the AACC substrate correlate with increases in rate constants for hydrolysis of the individual DNA strand compared to the wild-type protein. It is possible that the aspartate (D) better accommodates coordination of divalent metal ions close to the scissile phosphate of the bottom-strand on DNA substrates where central 4 bases cause perturbations in DNA structure that cannot be accommodated by the wild-type glutamate (E). The diversity of structural changes observed with different meganuclease–substrate complexes indicates that there is no common structural perturbation that can globally explain differences in cleavage efficiency (16,18). However, our data show that specific combinations of active site residues are better suited for indirect readout of DNA structures with different central 4 sequences, and that the shape of the active site rather than strict residue identity is a critical factor in coordination of divalent metal ion for efficient cleavage (58).
Implications for manipulating cleavage preference of meganucleases
Rationale engineering of protein-DNA specificity is aided by a protein-DNA code, best exemplified by transcription activator-like effectors and to a lesser extent by zinc-finger domains (reviewed in (11)). Current data are insufficient to determine if a meganuclease active site-central 4 base code exists because the number of target sites where the orientation of the meganuclease on the central 4 bases has been characterized is small, and most target sites are A/T rich (59). This low sequence diversity limits the accuracy of protein-DNA covariation predictions. Moreover, indirect readout of DNA structure and flexibility is composed of multiple molecular interactions that are difficult to ascribe to a single amino acid-base contact. This issue is particularly evident when considering the roles of residues that bind divalent metal ions for DNA cleavage and binding. However, there is precedent that changing the identity of metal-coordinating residues impacts cleavage fidelity. For instance, in the type II restriction enzyme KpnI, a D148E substitution in the active site generates an enzyme with higher cleavage fidelity than the wild-type enzyme (60,61), paralleling our observation that the opposite substitution (E to D at position 184) generates I-LtrI and I-OnuI variants with reduced fidelity. Some meganucleases naturally possess broad central 4 cleavage profiles, including I-PanMI where the catalytic network is AEGD (16). Inclusion of our strategy to meganuclease engineering workflows would help to identify active site variants that rescue activity on substrates not cleaved by the wild-type meganuclease, to fine-tune cleavage activity and specificity on select substrates and to select for variants with activity against novel central 4 combinations that would serve as starting scaffolds for further improvements. Given that a large proportion of restriction enzymes belong to the PD-(E/D)-XK superfamily (62,63), it may be possible that directed evolution of active site residues represents a new approach to modulate in vivo cleavage preference. This approach may be particularly relevant in enzymes where DNA binding and cleavage are uncoupled events (64,65).
Generalization and specialization of meganuclease function by toggling of active site resdiues
Natural selection shapes the specificity and activity of proteins as related to their cellular function. Some proteins have promiscuous or indiscriminate substrate specificity and catalyze the same reaction on a broad range of substrates with similar efficiency, whereas other proteins have narrow substrate specificity (66). This may be because the active sites and catalytic mechanisms of indiscriminate enzymes are more permissive to binding a wide range of substrate conformations, and subtle adaptations in active site structure through amino acid substitutions result in significant improvements in catalytic efficiency on suboptimal substrates (67–69). An interesting question is whether proteins that use indirect DNA readout are highly evolvable in the sense that relatively few amino acid changes would be required to modulate specificity or, to evolve new specificity altogether.
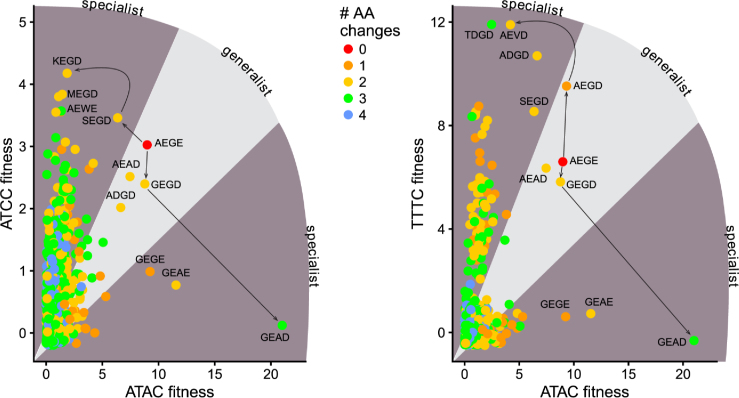
One unexpected finding from our profiling of active site variants was that relatively few amino acid substitutions in the active site change meganuclease cleavage preference from broad (or generalist) to narrow (or specialist) on a variety of substrates (Figure 10) (70). Moreover, residue combinations that show negative epistasis on one substrate often have positive epistasis on another substrate (the GEAE variant, for instance). From a biological perspective, the adaptability of the meganuclease active site correlates with the proposed cyclical lifecycle of meganucleases and other mobile endonucleases, characterized by initial invasion, continued spread and maintenance, and eventual degeneration and deletion (71,72). The ability to modulate cleavage preference by simple amino acid substitutions (73) would allow for meganucleases, at any point in the life cycle, the opportunity to expand to new target populations (the AEGD variant), to fine-tune activity for particular substrates (the GEAD variant) or to downregulate activity and increase specificity to avoid cellular toxicity (the SEGE variant), all of which would promote evolutionary persistence. Indeed, the AEGD variant enhances activity on the set of central 4 substrates that correspond to natural variation at both the I-OnuI and I-LtrI target sites in the rps3 gene (26) (Supplementary Figure S10).
Figure 10.
Evolutionary pathways for generalist or specialist enzymes. Potential evolutionary pathways showing how I-LtrI can transition from generalist to specialist cleavage preference on the cognate ATAC substrate to the ATCC or TTTC substrates. Plots are of I-LtrI variants with fitness values >6 for the indicated substrates, colored by number of amino acid substitutions in the co-evolving network relative to the wild-type AEGE network. Lines connect one evolutionary pathway.
Why then are these variants not widely spread amongst known meganucleases? It is possible that in a native context, these substitutions impart cellular toxicity or have other sub-optimal enzymatic activity. Alternatively, these variants may represent biophysically unstable evolutionary intermediates permitting expanded or contracted substrate ranges between more stable active site residue combinations. Although we did not measure thermal stability of variants identified here, we previously found no significant differences in experimentally determined melting temperatures for five I-LtrI network variants, or from computationally predictions of stability (23), implying that substitutions in active site residues do not dramatically affect protein stability.
In conclusion, our study demonstrates that profiling of active site fitness in combination with multiple DNA substrates can reveal insights into how meganucleases regulate cleavage preference. This data has implications for understanding evolutionary pathways to innovation of substrate specificity, as well as practical implications for customizing cleavage preference of re-purposed meganucleases for genome-editing applications. In principle, this strategy could be applied to other site-specific endonucleases.
DATA AVAILABILITY
Atomic coordinates and structure factors for I-LtrI structures have been deposited in the wwPDB under the following accession codes: I-LtrI AEGE-ATAC: 6BCE; I-LtrI AEAE-ATAC: 6BCF; I-LtrI GEGE-ATAC: 6BCG; I-LtrI AEAE-AACC: 6BCI; I-LtrI AEGD-AACC: 6BCT; I-LtrI AEGD-ATAC: 6BCN. Count tables and ALDEx2 outputs derived from deep sequencing for Figures 2–6 are available upon request from the authors. Illumina sequence data for the in vivo directed evolution experiments and in vitro cleavage specificity experiments have been deposited in the NCBI Short Read Archive under the accession code PRJNA491840.
Supplementary Material
ACKNOWLEDGEMENTS
The Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptam-Woodward Medical Research Institute. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract number DE AC02 06CH 11357.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant [RPGIN-2015-04800 to D.R.E., RGPIN-2015-03878 to G.G., RGPIN-2015-06658 to G.H.]; Canadian Institutes of Health Research [MOP-89903 to M.J.]; NSERC Canada Graduate Scholarship ( to T.M.). Funding for open access charge: NSERC Discovery Grant [RPGIN-2015-04800 to D.R.E.]., and internal University of Western Ontario funding (to D.R.E.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Belfort M., Derbyshire V., Parker M.M., Cousineau B., Lambowitz A.M.. Craig N.L., et al. Mobile introns: pathways and proteins. Mobile DNA II. 2002; Washington, D.C.: American Society of Microbiology; 761–783. [Google Scholar]
- 2. Stoddard B.L. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mobile DNA. 2014; 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grishin A., Fonfara I., Alexeevski D., Spirin S., Zanegina O., Karyagina A., Alexeyevsky D., Wende W.. Identification of conserved features of LAGLIDADG homing endonucleases. J. Bioinform. Comput. Biol. 2010; 08:453–469. [DOI] [PubMed] [Google Scholar]
- 4. Dalgaard J.Z., Klar A.J., Moser M.J., Holley W.R., Chatterjee A., Saira Mian I.. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 1997; 25:4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jurica M.S., Monnat R.J. Jr, Stoddard B.L.. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-Cre I. Mol. Cell. 1998; 2:469–476. [DOI] [PubMed] [Google Scholar]
- 6. Wang L., Smith J., Breton C., Clark P., Zhang J., Ying L., Che Y., Lape J., Bell P., Calcedo R. et al. . Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat. Biotechnol. 2018; 8:717–725. [DOI] [PubMed] [Google Scholar]
- 7. Redondo P., Prieto J., Munoz I.G., Alibes A., Stricher F., Serrano L., Cabaniols J.P., Daboussi F., Arnould S., Perez C. et al. . Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008; 456:107–111. [DOI] [PubMed] [Google Scholar]
- 8. Takeuchi R., Choi M., Stoddard B.L.. Redesign of extensive protein-DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4061–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arnould S., Perez C., Cabaniols J.-P., Smith J., Gouble A., Grizot S., Epinat J.-C., Duclert A., Duchateau P., Pâques F.. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007; 371:49–65. [DOI] [PubMed] [Google Scholar]
- 10. Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. et al. . Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Trans. Med. 2015; 7:307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bogdanove A.J., Bohm A., Miller J.C., Morgan R.D., Stoddard B.L.. Engineering altered protein–DNA recognition specificity. Nucleic Acids Res. 2018; 46:4845–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silva G.H., Belfort M.. Analysis of the LAGLIDADG interface of the monomeric homing endonuclease I-DmoI. Nucleic Acids Res. 2004; 32:3156–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stoddard B.L. Homing endonuclease structure and function. Q. Rev. Biophys. 2005; 38:49–95. [DOI] [PubMed] [Google Scholar]
- 14. Arnould S., Chames P., Perez C., Lacroix E., Duclert A., Epinat J.-C., Stricher F., Petit A.-S., Patin A., Guillier S. et al. . Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006; 355:443–458. [DOI] [PubMed] [Google Scholar]
- 15. Seligman L.M., Chisholm K.M., Chevalier B.S., Chadsey M.S., Edwards S.T., Savage J.H., Veillet A.L.. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002; 30:3870–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambert A.R., Hallinan J.P., Shen B.W., Chik J.K., Bolduc J.M., Kulshina N., Robins L.I., Kaiser B.K., Jarjour J., Havens K. et al. . Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity. Structure. 2016; 24:862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prieto J., Redondo P., López-Méndez B., DAbramo M., Merino N., Blanco F.J., Duchateau P., Montoya G., Molina R.. Understanding the indirect DNA read-out specificity of I-CreI Meganuclease. Sci. Rep. 2018; 8:10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Molina R., Redondo P., Stella S., Marenchino M., D’Abramo M., Gervasio F.L., Charles Epinat J., Valton J., Grizot S., Duchateau P. et al. . Non-specific protein-DNA interactions control I-CreI target binding and cleavage. Nucleic Acids Res. 2012; 40:6936–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takeuchi R., Lambert A.R., Mak A.N., Jacoby K., Dickson R.J., Gloor G.B., Scharenberg A.M., Edgell D.R., Stoddard B.L.. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13077–13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jarjour J., West-Foyle H., Certo M.T., Hubert C.G., Doyle L., Getz M.M., Stoddard B.L., Scharenberg A.M.. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009; 37:6871–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scalley-Kim M., McConnell-Smith A., Stoddard B.L.. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007; 372:1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takeuchi R., Certo M., Caprara M.G., Scharenberg A.M., Stoddard B.L.. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009; 37:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McMurrough T.A., Dickson R.J., Thibert S.M., Gloor G.B., Edgell D.R.. Control of catalytic efficiency by a coevolving network of catalytic and noncatalytic residues. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2376–E2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grizot S., Smith J., Daboussi F., Prieto J., Redondo P., Merino N., Villate M., Thomas S., Lemaire L., Montoya G. et al. . Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009; 37:5405–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silva G.H., Belfort M.. Analysis of the LAGLIDADG interface of the monomeric homing endonuclease I-DmoI. Nucleic Acids Res. 2004; 32:3156–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sethuraman J., Majer A., Friedrich N.C., Edgell D.R., Hausner G.. Genes within Genes: multiple LAGLIDADG homing endonucleases target the ribosomal protein S3 gene encoded within an rnl Group I intron of ophiostoma and related taxa. Mol. Biol. Evol. 2009; 26:2299–2315. [DOI] [PubMed] [Google Scholar]
- 27. Chan Y.-S., Takeuchi R., Jarjour J., Huen D.S., Stoddard B.L., Russell S.. The design and in vivo evaluation of engineered I-OnuI-based enzymes for HEG gene drive. PloS One. 2013; 8:e74254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolfs J.M., DaSilva M., Meister S.E., Wang X., Schild-Poulter C., Edgell D.R.. MegaTevs: single-chain dual nucleases for efficient gene disruption. Nucleic Acids Res. 2014; 42:8816–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen Z. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 2005; 33:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collaborative C.P. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 31. Adams P.D., Grosse-Kunstleve R.W., Hung L.-W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 32. Lu X.-J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Halford S.E., Johnson N.P., Grinsted J.. The EcoRI restriction endonuclease with bacteriophage λ DNA. Kinetic studies. Biochem. J. 1980; 191:581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gloor G.B., Hummelen R., Mackliam M.J., Dickson R.J., Fernandes A.D., MacPhee R., Reid G.. Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS ONE. 2010; 5:e15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fernandes A.D., Reid J.N., Macklaim J.M., McMurrough T.A., Edgell D.R., Gloor G.B.. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014; 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernandes A.D., Macklaim J.M., Linn T.G., Reid G., Gloor G.B.. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS One. 2013; 8:e67019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aitchison J. The Statistical Analysis of Compositional Data. 1986; London: Chapman & Hall. [Google Scholar]
- 38. Van den Boogaart K.G., Tolosana-Delgado R.. Analyzing compositional data with R. 2013; London: Springer. [Google Scholar]
- 39. Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J.. Microbiome datasets are Compositional: And this is not optional. Front. Microbiol. 2017; 8:2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. 1995; 57:289–300. [Google Scholar]
- 41. Thorsen J., Brejnrod A., Mortensen M., Rasmussen M.A., Stokholm J., Al-Soud W.A., Sørensen S., Bisgaard H., Waage J.. Large-scale benchmarking reveals false discoveries and count transformation sensitivity in 16S rRNA gene amplicon data analysis methods used in microbiome studies. Microbiome. 2016; 4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gloor G.B., Macklaim J.M., Fernandes A.D.. Displaying variation in large Datasets: Plotting a visual summary of effect sizes. J. Comput. Graph. Stat. 2016; 25:971–979. [Google Scholar]
- 43. Hawinkel S., Mattiello F., Bijnens L., Thas O.. A broken promise: microbiome differential abundance methods do not control the false discovery rate. Brief. Bioinform. 2017; doi:10.1093/bib/bbx104. [DOI] [PubMed] [Google Scholar]
- 44. Quinn T.P., Erb I., Richardson M.F., Crowley T.M. Understanding sequencing data as compositions: an outlook and review. Bioinformatics. 2018; 44:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakagawa S., Cuthill I.C.. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol. Rev. Camb. Philos. Soc. 2007; 82:591–605. [DOI] [PubMed] [Google Scholar]
- 46. Sullivan G.M., Feinn R.. Using effect Size-or why the P value is not enough. J. Grad. Med. Educ. 2012; 4:279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Macklaim J.M., Fernandes A.D., Di Bella J.M., Hammond J.-A., Reid G., Gloor G.B.. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome. 2013; 1:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wickham H. ggplot2: elegant graphics for data analysis. 2009; NY: Springer. [Google Scholar]
- 49. Schneider T.D., Stephens R.M.. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990; 18:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niu Y., Tenney K., Li H., Gimble F.S.. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J. Mol. Biol. 2008; 382:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Babic A.C., Little E.J., Manohar V.M., Bitinaite J., Horton N.C.. DNA distortion and specificity in a Sequence-Specific endonuclease. J. Mol. Biol. 2008; 383:186–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Joshi H.K., Etzkorn C., Chatwell L., Bitinaite J., Horton N.C.. Alteration of sequence specificity of the type II restriction endonuclease HincII through an indirect readout mechanism. J. Biol. Chem. 2006; 281:23852–23869. [DOI] [PubMed] [Google Scholar]
- 53. Martin A.M., Sam M.D., Reich N.O., Perona J.J.. Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease. Nat. Struct. Biol. 1999; 6:269–277. [DOI] [PubMed] [Google Scholar]
- 54. Little E.J., Babic A.C., Horton N.C.. Early interrogation and recognition of DNA sequence by indirect readout. Structure. 2008; 16:1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B.. The role of DNA shape in protein–DNA recognition. Nature. 2009; 461:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Koudelka G.B., Mauro S.A., Ciubotaru M.. Indirect readout of DNA sequence by Proteins: the roles of DNA Sequence-Dependent intrinsic and extrinsic forces. Prog. Nucleic Acid Res. Mol. Biol. 2006; 81:143–177. [DOI] [PubMed] [Google Scholar]
- 57. Lawson C.L., Berman H.M.. Rice PA, Correll CC. Indirect readout of DNA sequence by proteins. Protein-Nucleic Acid Interactions: Structural Biology. 2008; Cambridge: Royal Society of Chemistry; 66–90. [Google Scholar]
- 58. Skirgaila R., Grazulis S., Bozic D., Huber R., Siksnys V.. Structure-based redesign of the catalytic/metal binding site of Cfr10I restriction endonuclease reveals importance of spatial rather than sequence conservation of active centre residues. J. Mol. Biol. 1998; 279:473–481. [DOI] [PubMed] [Google Scholar]
- 59. Taylor G.K., Petrucci L.H., Lambert A.R., Baxter S.K., Jarjour J., Stoddard B.L.. LAHEDES: the LAGLIDADG homing endonuclease database and engineering server. Nucleic Acids Res. 2012; 40:W110–W116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vasu K., Nagamalleswari E., Zahran M., Imhof P., Xu S.-Y., Zhu Z., Chan S.-H., Nagaraja V.. Increasing cleavage specificity and activity of restriction endonuclease KpnI. Nucleic Acids Res. 2013; 41:9812–9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saravanan M., Vasu K., Nagaraja V.. Evolution of sequence specificity in a restriction endonuclease by a point mutation. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10344–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kosinski J., Feder M., Bujnicki J.M.. The PD-(D/E)-XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC bioinformatics. 2005; 6:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pingoud A., Wilson G.G., Wende W.. Type II restriction endonucleases a historical perspective and more. Nucleic Acids Res. 2014; 42:7489–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gimble F.S., Stephens B.W.. Substitutions in conserved dodecapeptide motifs that uncouple the DNA binding and DNA cleavage activities of PI-SceI endonuclease. J. Biol. Chem. 1995; 270:5849–5856. [DOI] [PubMed] [Google Scholar]
- 65. Waugh D.S., Sauer R.T.. Single amino acid substitutions uncouple the DNA binding and strand scission activities of Fok I endonuclease. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:9596–9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Copley S.D. Shining a light on enzyme promiscuity. Curr. Opin. Struct. Biol. 2017; 47:167–175. [DOI] [PubMed] [Google Scholar]
- 67. Ben-David M., Elias M., Filippi J.-J., Duñach E., Silman I., Sussman J.L., Tawfik D.S.. Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1. J. Mol. Biol. 2012; 418:181–196. [DOI] [PubMed] [Google Scholar]
- 68. Ben-David M., Wieczorek G., Elias M., Silman I., Sussman J.L., Tawfik D.S.. Catalytic metal ion rearrangements underline promiscuity and evolvability of a metalloenzyme. J. Mol. Biol. 2013; 425:1–11. [DOI] [PubMed] [Google Scholar]
- 69. Yang G., Hong N., Baier F., Jackson C.J., Tokuriki N.. Conformational tinkering drives evolution of a promiscuous activity through indirect mutational effects. Biochemistry. 2016; 55:4583–4593. [DOI] [PubMed] [Google Scholar]
- 70. Tawfik O.K. S D. Enzyme Promiscuity: a mechanistic and evolutionary perspective. Ann. Rev. Biochem. 2010; 79:471–505. [DOI] [PubMed] [Google Scholar]
- 71. Koufopanou V., Goddard M.R., Burt A.. Adaptation for horizontal transfer in a homing endonuclease. Mol. Biol. Evol. 2002; 19:239–246. [DOI] [PubMed] [Google Scholar]
- 72. Goddard M.R., Burt A.. Recurrent invasion and extinction of a selfish gene. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:13880–13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Roy A.C., Wilson G.G., Edgell D.R.. Perpetuating the homing endonuclease life cycle: identification of mutations that modulate and change I-TevI cleavage preference. Nucleic Acids Res. 2016; 44:7350–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for I-LtrI structures have been deposited in the wwPDB under the following accession codes: I-LtrI AEGE-ATAC: 6BCE; I-LtrI AEAE-ATAC: 6BCF; I-LtrI GEGE-ATAC: 6BCG; I-LtrI AEAE-AACC: 6BCI; I-LtrI AEGD-AACC: 6BCT; I-LtrI AEGD-ATAC: 6BCN. Count tables and ALDEx2 outputs derived from deep sequencing for Figures 2–6 are available upon request from the authors. Illumina sequence data for the in vivo directed evolution experiments and in vitro cleavage specificity experiments have been deposited in the NCBI Short Read Archive under the accession code PRJNA491840.