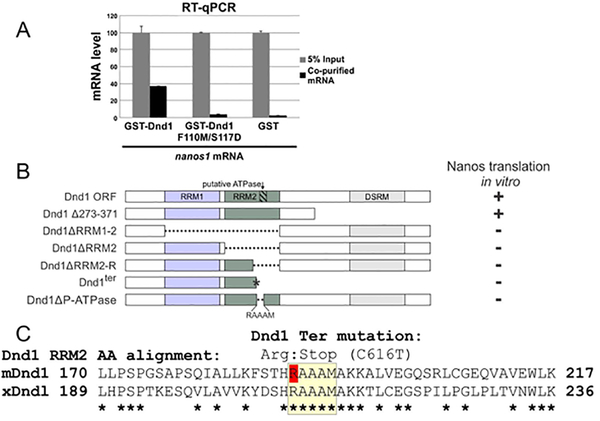
Figure 1. Dnd1 RRM1 and RRM2 are required for binding and translation of nanos1 RNA respectively.
A. RRM1 of Dnd1 protein is required to bind to nanos1 mRNA. mRNAs were pulled down by recombinant GST-Dnd1 or GST-Dnd1-F110M/S117D proteins in an in vitro RIP assay and bound mRNA measured by qPCR. Ratio between the pulldown mRNAs and 5% of mRNA input is shown. GST protein served as a negative control. Data are shown as mean ± s.d. **p<0.01 of two independent experiments. B. Deletion mapping analysis of Dnd1 protein shows that the right half region of RRM2 (RRM2-R, Δ198–237) is the smallest region required to promote nanos1 translation. The putative ATPase site (RAAAM) lies within the RRM2-R region. Analysis was based on at least two independent in vitro translation experiments using wheat germ extracts. Findings were confirmed in vivo using oocytes from two different females. Raw data is shown in SFig.1. C. Alignment of the RRM2 right half region of mouse and Xenopus Dnd1. Putative ATPase region is highlighted in yellow. Note: the point mutation C616T that replaces an arginine creates a stop codon in the mouse Dnd1 ter mutation (red R). Asterisks indicate identity (Clustal Omega, EMBL-EBI, UK).