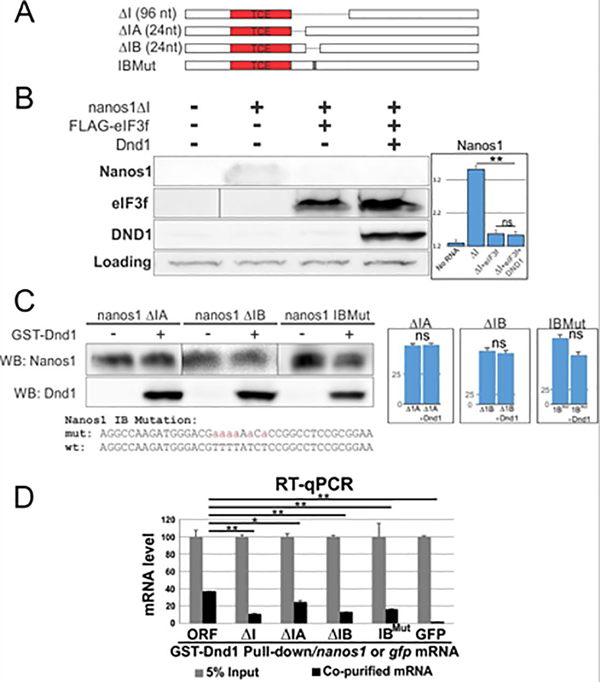
Figure 8. Dnd1 binds to TCE adjacent U-rich region of nanos1 mRNA.
A. Diagram shows serial deletions of nanos1 mRNA used in in vitro translation and RIP assays. nanos1 ΔI region is missing 96 nt downstream of the TCE. Smaller deletions within region I were carried out (ΔIA and ΔIB, 24 nt each). B. WG extracts were supplemented with either 500 pg of nanos1 mRNA with or without 1ug of translation repressor eIF3f and 100 pg of xDnd1 protein and analyzed by western blot. A non-specific band served as a loading control. The presence of Dnd1 protein did not promote the translation of nanos1 mRNA missing region I (ΔI) after repression by eIF3f. Quantitation of blot shown on right. C. Wheat germ extracts with either 500 pg of nanos1ΔIA/B or nanos1 region IB with mutations in U-rich sequence. mRNAs were incubated with or without Dnd1 protein and analyzed by western blotting. Dnd1 does not improve nanos1 translation. Quantitation of blot shown on right. Experiments repeated twice. D. RIP analysis. Different nanos1 mRNAs with deletion or mutation were pulled down by GST-Dnd1 and measured by qPCR. Ratio between the pulldown and 5% of RNA input is shown. GFP served as a negative control. All experiments were repeated at least twice. Quantification of results shown in B, C are shown as mean ± s.d. Two-tailed t-tests were performed. ** P<0.01; * P<0.05; ns: no significant difference.