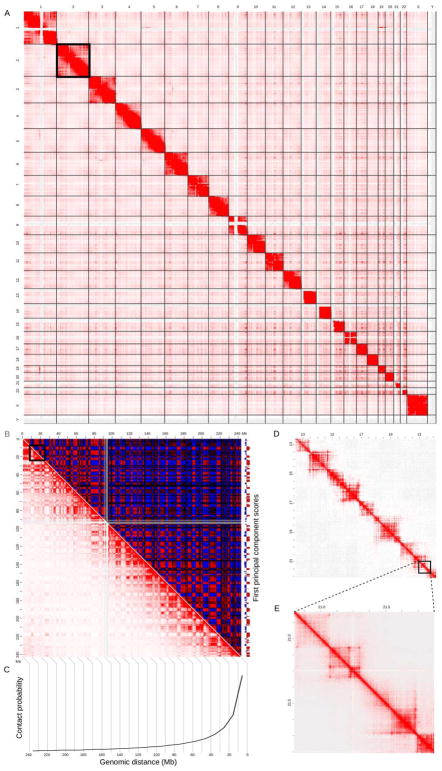
Figure 2. The iterative process of experimentation, hypothesis generation and confirmation of 3D genomic features, exemplified by the development of the loop extrusion model.
A) A Hi-C experiment results in the production of a contact matrix, which is visualized using a heat map. Computational analysis facilitates the detection of 3D genomic features such as topologically associating domains (TADs; white lines) and loops (white circles). Observations of such 3D features, aided by integration of linear genomic features (not shown), result in the development of a hypothesis as to their origin. B) A schematic representation of the loop extrusion model (described in the text), a hypothesis explaining the formation of TADs and loops. C) A polymer model of the chromosome is built based on a hypothesis such as the loop extrusion model. Simulations of the polymer model then produce an ensemble of 3D structures (only three are shown here) of the chromosome. The boundary element are highlighted as orange spheres while the rest of the chromosome is in blue. D) The ensemble of structures from loop extrusion model simulations are used to compute a contact map (Sanborn et al., 2015). This contact map is then compared to observed data (see A) leading to refinement of the hypothesis (see B) and/or details regarding its implementation, such as optimizing parameters (see C). Additionally, functional experiments can also be conducted to validate the hypothesis, potentially necessitating the generation and testing of new ones.