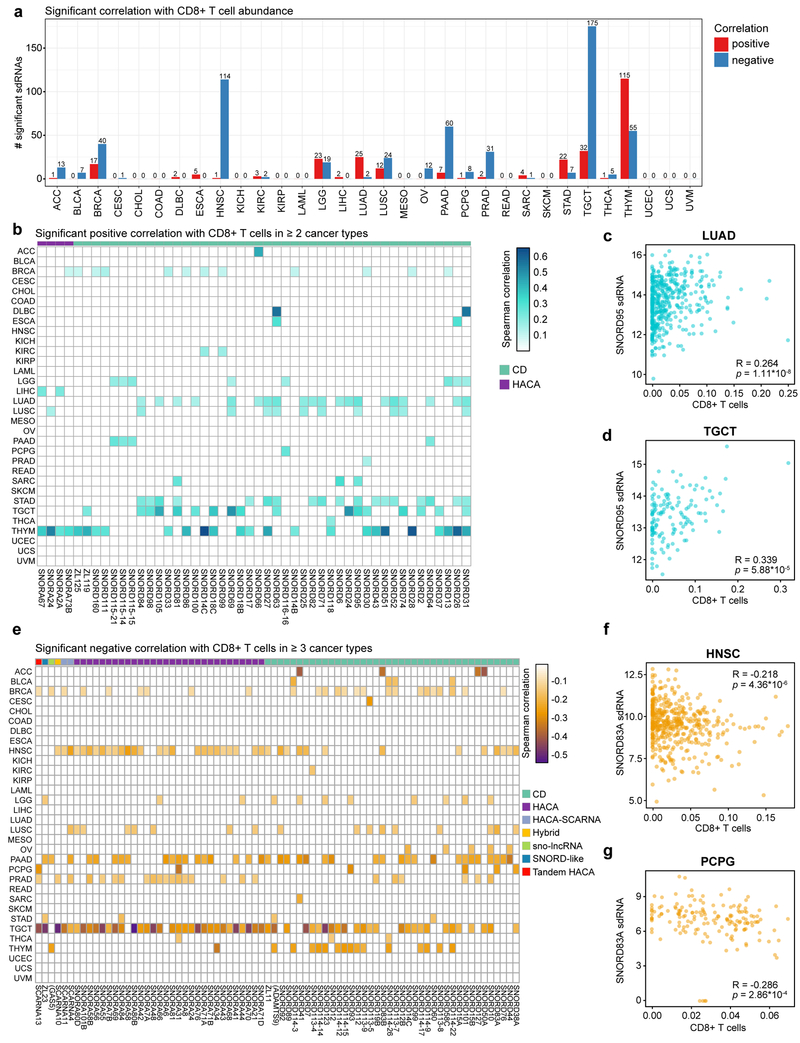
Figure 5: sdRNAs are correlated with CD8+ T cell infiltration in diverse cancers.
a. Bar plot depicting the number of significant sdRNAs in each cancer type, aggregated by parental snoRNA, in relation to CD8+ T cell abundance. Red, positive correlation; blue, negative correlation.
b. Heat map of sdRNAs positively correlated with CD8+ T cell abundance (adjusted p < 0.05, adjusted within each cancer type). For visibility, only sdRNAs that were positively correlated in two or more cancer types are shown. Boxes are colored according to the Spearman correlation with CD8+ T cell abundance. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
c. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD95 sdRNA expression in lung adenocarcinoma (LUAD, n = 454). R = 0.264, p = 1.11 * 10-8.
d. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD95 sdRNA expression in testicular germ cell tumors (TGCT, n = 135). R = 0.339, p = 5.88 * 10-5.
e. Heat map of sdRNAs negatively correlated with CD8+ T cell abundance (adjusted p < 0.05, adjusted within each cancer type). For visibility, only sdRNAs that were positively correlated in four or more cancer types are shown. Boxes are colored according to the Spearman correlation with CD8+ T cell abundance. Parental snoRNAs without annotated names are instead labeled by their host gene in parentheses. SnoRNA classifications are annotated on top based on a color legend on the right panel.
f. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD83A sdRNA expression in head and neck squamous cell carcinoma (HNSC, n = 435). R = −0.218, p = 4.36 * 10-6.
g. Scatter plot depicting the correlation between CD8+ T cell abundance and SNORD83A sdRNA expression in pheochromocytomas and paragangliomas (PCPG, n = 157). R = −0.286, p = 2.86 * 10-4.
c, d, f, g - Data are shown as tpm for smRNA-seq datasets.