Figure 7.
Changes in FLI1 Transgene Expression Dictates Forward Programming Outcome
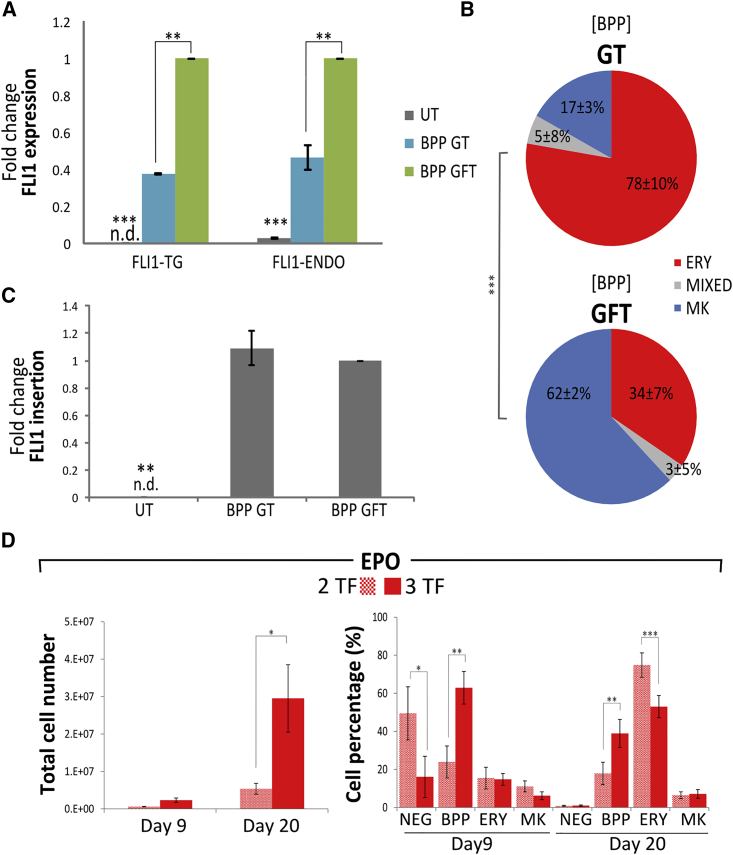
Day 9 CD41+ CD235a+ BPPs were sorted by flow cytometry into GATA1+/FLI1+/TAL1+ (GFT) and GATA1+/FLI1−/TAL1+ (GT) populations.
(A) FLI1 transgene and endogenous expression was measured by qRT-PCR on sorted populations and normalized to the GFT population; GFT (green bars), GT (blue bars), and untransduced (gray bars).
(B) The clonogenic potential of the GFT and GT-sorted BPPs cells was tested by CFU assay. The pie charts show colony distribution from both populations after 14 days (n = 2, mean%); erythroid colonies (ERY, red), megakaryocyte-erythrocyte mixed colonies (MIXED, gray), and megakaryocyte colonies (blue). Poisson regression analysis shows that the distribution of the number of colonies per cell type depends strongly on the set of expressed TFs (∗∗∗p < 2.2 × 10−16).
(C) Detection of integrated FLI1 provirus by qPCR in the genomic DNA of day 9 GFT and GT-sorted BPPs; UT, untransduced cells (n = 2, mean% normalized to GFT cells).
(D) Forward programming was performed using 3TFs (GATA1, FLI1, and TAL1) or 2TFs (GATA1 and TAL1) in EPO culture conditions. Cells were monitored by flow cytometry at days 9 and 20 post-transduction to determine viable cell numbers (left) and the CD41a/CD235a cell phenotype (right) (n = 5, mean% ± SEM). Cell numbers were significantly decreased with 2TFs as well the overall proportion of BPPs generated at days 9 and 20.